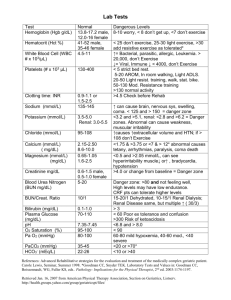
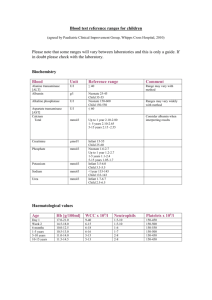
CVR-2010-389R1 Supplementary Material Methods 1. Preparation
advertisement

CVR-2010-389R1 Supplementary Material Methods 1. Preparation of neonatal rat cardiac fibroblast cultures Cardiac fibroblasts were prepared from hearts of newborn (0–2 day old) Sprague–Dawley rat pups, as described.1 This procedure yielded cultures that were >98% fibroblasts, as confirmed by negative immunofluorescent staining with mouse desmin monoclonal antibody. Briefly, freshly harvested CFs were grown to confluence (3 days) in Dulbecco’s modified minimum essential medium (DMEM) with 10% newborn calf serum. CFs were then trypsinized (TrypLETM Express, Gibco®) and replated on deformable membranes coated with collagen-IV [1 µg/cm2]) on Bioflex® plates (Flexcell International Corp, Hillsborough, NC), at a density of 0.45 × 106 cells/well in DMEM/M199 medium and maintained at 37°C in humid air with 5% CO2. Collagen IV has been shown to be primarily bound by α1β1 integrin.2 Since in our study β1 integrin involved as a mechanical signal transducer, we chosen collagen IV as our matrix for CFs. Additionally, the cardiac fibroblasts readily attach to collagen IV and undergo minimal migration. The medium was changed to serum-free DMEM/M199 and experiments were performed 24 h later. For all the stretch experiments, cells were exposed to 20% equiaxial stretch. 2. Adenovirus expansion Amplification of adenoviruses was performed in transformed 293 human embryonic kidney (HEK) cells (CRL-1573, American Type Culture Collection, Manassas, VA), followed by purification on cesium chloride gradients.3 The multiplicity of viral infection (MOI) was determined for each adenovirus by dilution assay in HEK-293 cells. CFs were titrated with viruses were titrated to maximize expressed protein, but prevent viral toxicity and detachment of 1 CVR-2010-389R1 cells from deformable membranes during stretch procedures. Expression of the target proteins was determined using flow cytometry and Western blot analysis. The shape and cell areas of CFs expressing tac-β1, as well as dominant negative and constitutively Rho GTPases were similar compared to control cells (non-infected and virus controls). However, the cell borders of CFs expressing tac-β1 had a slight “smooth” appearance consistent with partial inhibition of 1 integrin detachment to extracellular matrix. 3. Western blot analysis Cells were scrapped into freshly prepared cell lysis buffer (20 mmol/L Tris-HCl [pH 7.5], 150 mmol/L NaCl, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L sodium orthovanadate, 1 μg/ml leupeptin, 10 μg/ml aprotinin, 10 mmol/L 2aminoethyl)-benzenesulfonyl fluoride hydrochloride, 1 mmol/L ethylenediaminetetraacetic acid [EDTA], 1 mmol/L ethylene glycol tetraacetic acid [EGTA], 1% Triton® X-100) to solubilize structure and membrane bound proteins. Western blot analysis was performed as previously described.1 Briefly, equal amounts of protein (30 μg) from cell lysates were separated by 10-12% SDS-PAGE and blotted onto polyvinylidene fluoride (PVDF, GE Healthcare) transfer membranes. Blots were blocked for 2 h with 5% protease-free bovine serum albumin [BSA] in TBST buffer (10 mmol/L Tris [pH 7.6], 100 mmol/L NaCl, 0.1% Tween® 20). Blots were incubated with the primary antibodies in 5% BSA in TBST buffer overnight at 40C with light agitation. Following incubation with primary antibody, blots were washed with TBST buffer and incubated with appropriate horseradish peroxidaselabeled secondary antibodies in 1% BSA in TBST buffer for 1 h at 37°C. After blots were washed with TBST, target proteins were detected using enhanced chemiluminescence (ECL). Densities of bands were determined using ImageQuant software (GE Healthcare). Signals 2 CVR-2010-389R1 from phosphoproteins were normalized to the total protein, obtained by stripping and reprobing blots with the corresponding total antibody. Blots were again stripped and probed with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (sc-25778, Santa Cruz Biotechnology Inc., Santa Cruz, CA) to confirm equal loading. The bands of interest were quantified using scanning densitometry. 4. Rho GTPase pull down assays Rho GTPases cycle from inactive (GDP-bound) to active (GTP-bound) forms that interact with and regulate components of intracellular signaling pathways. This cycle is under the control of several classes of regulatory protein. Under basal conditions, the Rho GTPases complex with the cytosolic GDP-dissociation inhibitor (GDI), which stabilizes the inactive GDP-bound form. Upon cell stimulation, the GTPase dissociates from the GDI and translocates to the plasma membrane. Subsequently the GTPase releases GDP and binds GTP, a reaction promoted by guanine nucleotide exchange factors (GEFs), which leads to the interaction with specific molecular targets. Only the active form of the GTPase interacts with downstream effectors. In this method we precipitate the active GTPase using glutathione S-transferase (GST) beads coated with the downstream effectors of respective GTPase. The affinity precipitation assay detects active Rac 1 by using p21-binding domain (PBD) of PAK 1 fused to GST to immunoprecipitate the active GTPase (Rac-GTP). The precipitated GTPase is then quantified by Western blotting, using specific antibodies (Fig. 1 & 4B). RhoA activation was measured in a similar fashion, using the Rhotkin binding domain (RBD). 5. Sepherose-glutathione S-transferase (GST) bead preparation L-broth medium (10 ml) containing ampicillin (100 µg/ml) was incubated with transformed 3 CVR-2010-389R1 bacterium (gift from Dr. Ugra Singh, South Carolina Medical School, Columbia, SC) and grown overnight at 37oC with constant stirring. The overnight culture was extended in 250 ml of Lbroth medium-ampicillin for 3 h at 370C with constant stirring followed by adding 0.8 mM isopropyl--D-thiogalactopyranoside for 2 h at 370C to increase bacterial transcription. The bacterial suspension was pelleted by centrifugation (6,000 g) for 15 min at 40C. The pellet was resuspended in 5 ml of bacterial lysis buffer (50 mmol/L Tris-HCl [pH 7.5], 150 mmol/L NaCl, 5 mmol/L MgCl2, 1 mmol/L dithiothreitol [DTT], 1 mmol/L EDTA, 0.1% Triton® X-100, 10 μmol/L phenylmethylsulfonyl fluoride [PMSF], 10 µg/ml aprotinin, 10 µg/mL leupeptin, 1 mg/ml lysozyme, 15 µg/ml DNase I) incubated for 1 h on ice, sonicated and centrifuged (20,000 g) at 40C for 20 min. The fusion protein was purified from the supernatant with previously swollen glutathione-sepherose beads (Sigma Chemical Company, St. Louis, MO) and washed in lysis buffer by incubation for 2 h at 40C. The bacterially produced GST fusion protein containing the Rac1 interactive binding domain of PAK, prebound to glutathione–sepharose beads (GST–PBD), and the RhoA interactive binding domain of Rhotekin, prebound to glutathione–sepharose beads (GST–RBD), were centrifuged (2,000 g) for 5 min, washed in bead wash buffer (25 mmol/L Tris-HCl [pH 7.4], 10 mmol/L NaCl, 1 mmol/L DTT, 1 mmol/L EDTA, 1% Triton® X-100, 10 μmol/L PMSF, 10 µg/mL aprotinin and 10 µg/mL leupeptin) and transferred to storage solution (50 mmol/L Tris-HCl [pH 7.5], 150 mmol/L NaCl, 5 mmol/L MgCl2, 10% glycerol, 0.5% Triton® X-100, 10 μmol/L PMSF, 10 µg/mL aprotinin, 10 µg/mL leupeptin) prior to use in experiments.4 6. Quantitative measurement of Ao mRNA using real-time RT-PCR A commercial kit (ToTALLY RNATM, Ambion, Austin, TX) was used to isolate total RNA from CFs. First strand cDNA was reverse-transcribed (RT) with MultiScribeTM reverse transcriptase 4 CVR-2010-389R1 and random hexamer primers using the High-Capacity cDNA Archive kit (ABI Prism, Foster City, CA) for RT-PCR. Real-time RT-PCR was carried out in a MX3005P (Stratagene, La Jolla, CA) thermocycler using Taqman® Universal PCR Master Mix (ABI, USA). Absolute levels of Ao mRNA were quantified using 21-base sense (5'-AGCACGACTTCCTGACTTGGA-3') and antisense primers (5'-TTGTAGGATCCCCGATTTCC-3'), which span the second intron of the genomic sequence and produce an 88 base-pair amplicon. The amplified Ao DNA was detected using oligonucleotide (6FAM-5'-CCGTCTGACCCTGCCGCAGC-3'-TAMRA), as the reporter probe. Parallel reactions, yeast RNA (carrier RNA) spiked with known amounts of synthetic Ao RNA, were used to convert sample determinations to absolute values. Ao primers and reporters were multiplexed with those for GAPDH, which was used as a "house-keeper" RNA (proprietary reagents supplied by ABI). Multiplexing was used to control for RNA loading, degradation and PCR efficiency. Figure Legends Figure S1. Stretch-induced temporal activation of stress-activated protein kinases p38 and JNK 1/2 in cardiac fibroblasts. (A) CFs were stretched (20%) for various times (2 minutes - 24 hours), after which cells were harvested. Cell lysates were denatured in Laemmli sample buffer, separated on 10% SDS-PAGE, transferred to PVDF membranes and immunoblotted, as described in the above Supplementary methods (Section 3). (B) Bar graphs show fold changes in p38 and JNK1/2 phosphorylation after stretch, as compared to no stretch. Results are means ± SEM from 4 independent experiments. CFs, Neonatal rat ventricular cardiac fibroblasts. References 1. Dostal DE, Booz GW, Baker KM. Regulation of angiotensinogen gene expression and 5 CVR-2010-389R1 protein in neonatal rat cardiac fibroblasts by glucocorticoid and beta-adrenergic stimulation. Basic Res Cardiol 2000;95:485-490. 2. Tulla M, Pentikäinen OT, Viitasalo T, Käpylä J, Impola U, Nykvist P et al., Selective binding of collagen subtypes by integrin alpha 1I, alpha 2I, and alpha 10I domains. J Biol Chem 2001;276:48206-48212. 3. Lal H, Verma SK, Smith M, Guleria RS, Lu G, Foster DM et al., Stretch-induced MAP kinase activation in cardiac myocytes: differential regulation through beta1-integrin and focal adhesion kinase. J Mol Cell Cardiol 2007;43:137-147. 4. Yang FC, Atkinson SJ, Gu Y, Borneo JB, Roberts AW, Zheng Y et al., Rac and Cdc42 GTPases control hematopoietic stem cell shape, adhesion, migration, and mobilization. Proc Natl Acad Sci U S A 2001;98:5614-5618. 6