Genetics Concept Activity: Molecular Pedigrees
advertisement

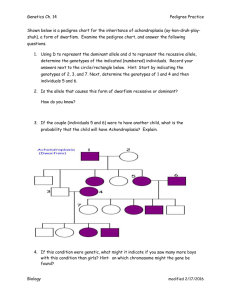
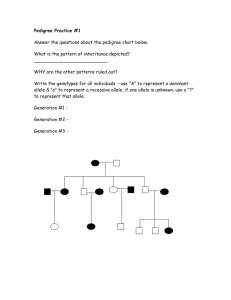
Investigative Laboratory: Generating Molecular Pedigrees Background Objective: To utilize your knowledge of agarose gel electrophoresis, DNA structure and pedigree analysis to identify a restriction fragment length polymorphism (RFLP) marker that is linked to your module mystery allele of interest. The marker fragment can then be used to track the allele’s presence or absence in each member of your module mystery family, and will help you to determine the associated trait’s mode of inheritance. Background: Pedigrees The types of controlled experimental crosses that we have used to determine the modes of inheritance in other species are simply not possible in human populations. Rather, human geneticists must infer genotypes and modes of inheritance using matings that have already happened. As your textbooks described, these matings can be represented graphically in a family pedigree. Symbols commonly used in pedigrees are shown in the table below. The information needed to fill out a human pedigree can be acquired in a variety of ways. You have already completed the first method: interviews with family members. If a genetic marker is available for a disease, it can also be used to predict whether or not an individual carries a disease gene. Restriction Fragment Length Polymorphisms (RFLP’s) Restriction endonucleases cut DNA at specific nucleotide sequences called restriction sites. Even a single base change to a restriction site can prevent cutting by a restriction enzyme (Figure 2). Figure 2. Effect of a single base mutation on restriction endonuclease function. A. Eco RI normally cuts at the DNA sequence GAATTC. Let’s say that Allele “A” has the following DNA sequence: ATTAACGTGAATTCCTTACCTATCGGTCGTAGCGTGCGGTGAATTCGCGGA + Eco RI ATTAACGTG AATTCCTTACCTATCGGTCGTAGCGTGCGGTGAATT CGCGGA When Eco RI cuts Allele “A”, it generates a 36 bp restriction fragment. AATTCCTTACCTATCGGTCGTAGCGTGCGGTGAATT B. If GAATTC is changed to GAGTTC at the second restriction site, Eco RI will not cut. Let’s say that Allele “a” has the following DNA sequence: ATTAACGTGAATTCCTTACCTATCGGTCGTAGCGTGCGGTGAG TTCGCGGA + Eco RI ATTAACGTG AATTCCTTACCTATCGGTCGTAGCGTGCGGTGAG TTCGCGGA When Eco RI cuts Allele “a”, it generates a 42 bp restriction fragment AATTCCTTACCTATCGGTCGTAGCGTGCGGTGAG TTCGCGGA C. Thus, every individual who carries the ‘A’ allele will have a 36 bp restriction fragment, and every individual who carries the ‘a’ allele will have a 42 bp restriction fragment. This difference in restriction fragment sizes is called a restriction fragment length polymorphism or RFLP. RFLPs can be used to determine the genotypes of every individual in a pedigree, as shown below, because each genotype has a unique restriction fragment pattern. AA aa Aa 42 bp 36 bp Use of RFLP’s as genetic markers Genetic markers are alleles that can be used to keep track of a particular gene in an individual or population. The marker is not the disease gene itself. Rather, it is a recognizable allele that segregates with the disease allele because they are very tightly linked on the same chromosome. Thus, they tend to be inherited together. In the Module Mystery Molecular Pedigree lab, you will try to determine which restriction fragment is segregating with the disease allele in your family. You will then use this information to generate a pedigree for that family that includes the genotypes of all individuals. That pedigree should be part of your final module mystery poster. First, however, you and your team will practice this approach in the next Team Application Problems assignment.