Exam I Key 2009

MBMB 451A Exam I September 24, 2009 Name: ___________________________
(1) Write out the shorthand notation for the nucleic acid structure displayed below (2).
Short hand notation is as follows:
A T C G
MBMB 451A Exam I September 24, 2009 Name: ___________________________
(2) Give the name of the following three compounds (2+2+2)
A
.
B. C.
A.
Adenine
B.
Adenosine
C.
Adenosine phosphate, adenosine monophosphate, adenylic acid, 5’adenylic acid, adenylate, 5’ adenylate,
(3) What are the key physical differences between A- and B-DNA? The A-DNA structure is representative of what kind of naturally occurring nucleic acid structure?
(2+2+1)
Key features: 6 angstrom hole in A DNA that is in the middle, no hole in B-DNA. Both are right handed. The major groove is wider in B-DNA than in A-DNA. Sugar pucker is also different between the two
Differences:
Parameters bp/turn twist/turn length/base
B-DNA
10bp
36°
0.34nm pitch helix diameter major groove minor groove sugar pucker glycosidic bond
3.4nm
2.0nm base plane tilt away from helix axis 6° orientation right handed wide and deep narrow and deep
C2’endo anti
A DNA structures occuring naturally:
RNA-RNA hybrids
RNA-DNA hybrids
A-DNA
11.6bp
31°
0.29nm
3.4nm
2.6nm
20°
right handed narrow and deep wide and shallow
C3’endo anti
MBMB 451A Exam I September 24, 2009 Name: ___________________________
(4) What is a G quartet and where is it naturally found in the cell? Describe the type of base pairing present. (2+1+2)
The G-rich overhanging ends of telomeres dimerize by formation of cyclic tetramers called G-quartets. In these structures, guanine forms strong hoogsteen type base pairs.
G quartets are found in the telomeres.
(5) Proteins that are able to bind specific DNA sequences are able to recognize their cognate sequence using a series of hydrophobic, hydrogen bond donor and acceptor groups present in the major groove. Using the structures below identify what are the functional groups for each base pair that would be use in this recognition process.
For the G-C base pair:
Hydrophobic group Donor Group
G
C
A
-
-
For the A-T base pair:
C4-amino
Hydrophobic group Donor Group
C6-amine
Acceptor Group
N7, C6-carbonyl
Acceptor Group
N7
MBMB 451A Exam I September 24, 2009 Name: ___________________________
T methyl group C4-carbonyl
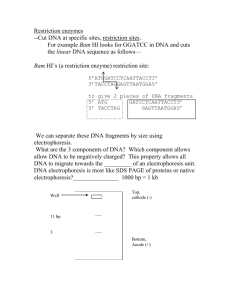
(6) What are the characteristics of type I, II, and III restriction endonucleases? (2+2+2)
TYPE I: single polypeptides, both endonuclease and methylase activities, cleave up to
1000bp from recognition site
MBMB 451A Exam I September 24, 2009 Name: ___________________________
TYPE II: separate endonuclease and methylase activities, cleave within recognition sequence
TYPE III: single polypeptides, both endonuclease and methylase activities, cleave ~25bp away
(7) How are samples generally prepared for measuring the levels of mRNA at a genomic level using a DNA microarray? Please describe all enzymes used in this process. Start by describing four different ways that cells can be lysed. The final step should be how the sample is applied to your microarray, treated and then visualized.(2+4+2)
Isolate total RNA from cells/organism
Anneal dT24 primer to poly A tail and extend using reverse transcriptase and dNTPs, this forms cDNAs from the mRNA population
Degrade RNA strand of DNA-RNA hybrid with RNase
Amplify with random primers using a DNA polymerase like Taq and fluorescently labeled nucleotides i. osmotic lysis - hypotonic solution, swelling and bursting ii. French press - high pressure & small orifice iii. sonicator iv. homogenizer -tissue grinder, v. glass beads versus mortar & pestle vi. dounce
The first step is to denature the cDNAs and then hybridized to the DNA microarray chip.
After adequate hybridization time, the DNA chip is washed and then scanned using a laser to measure the relative extent of fluorescence across the entire chip. By quantitating the fluorescent signals we can get an idea of the level of gene expression.
(8) Suppose that you want to examine the potential role of a particular gene on transcription at a genome level and use the expression profiling approach mentioned in
(7). What would be a key control for this experiment and would there be any differences in how you prepare your samples?
Controls: mRNA samples from WT strains
RNA from the WT and mutant samples are labeled with different dyes
Mix the cDNA probes from WT and mutants together and apply on microarray chips using the same protocol for hybridization described above
Scan chip separately for the two dyes and merge images.
Compare the different signal levels
MBMB 451A Exam I September 24, 2009 Name: ___________________________
(9) What is the difference between a strong and weak cation exchange resin? Why would one of these types of resin be preferred over the other?
Strong cation exchanger: stays charged across a large pH range. Advantage: pH does not affect ion exchange performances.
Weak cation exchanger: has a narrower pH range in which the functional group is charged.
Advantage: Can elute protein by changing pH. Disadvantage: need to more carefully monitor pH
Strong cation exchangers are preferred owing to their ability to work over a wide pH range
(10) Explain the basis of discontinuous gel electrophoresis to separate proteins by size and the principle of a “stacking” gel.
The resolution & focus of the protein bands is increased by using discontinuous gels
(Laemmli gels)- the stacking gel (pH 6.8, %T=3 to 5 %) & the resolving gel (pH 8.8,
%T= 5 to 20 %). %T represents acrylamide percentage. These gels are usually run at constant current. At pH=6.8, most of the glycine in the population exist as zwitterions with no negative charge (pKa 1 =2.45; pKa 2 =9.6; pI=6.025). At this stage, The glycine ions move slowly and tend to cause the proteins to pile up behind the high concentration of glycine.
Upon entering the resolving gel (pH=8.8), the glycine zwitterions deprotonate to the anionic form. The proportion of these ions increase from 0.0015% to 15.8%. Due to their small size, the glycine anions move very fast and leave the protein behind so they can migrate based on their own charge and size in an SDS-micelle.
(11) What is the composition of a nucleosome, chromatosome, 10 nm and 30 nm fibers?
Nucleosome = DNA + 2 each of H2A, H2B, H3 and H4
Chromatosome = nucleosome + H1
10nm fiber = beads on a string structure, polynucleosome chain, may or may not contain
H1
30nm fiber = solenoid or zig zag higher order form, highly ordered polynucleosome chain, generally contains H1.
(12) You digested chromatin from chicken erythrocyte nuclei for 20 minutes at 25 deg C with the following concentrations of DNAse I: 0, 0.1, 1.0 and 10 mg/ml. You loaded these samples along with molecular weight marker on a 6% polyacrylamide denaturing
MBMB 451A Exam I September 24, 2009 Name: ___________________________ gel. Below is shown a diagram of your gel. Unfortunately, you mixed up the samples and cannot be sure what was loaded in which lane of the gel. You are sure that molecular weight markers are loaded in the left lane, but you have forgotten the sizes of the marker bands.
(a) Which DNAse I concentration was used in each lane? Write your answers in the spaces provided on the left of the diagram.
(b) Provide approximate sizes for the bands in the marker lane (designated M). Write your answers in the spaces provided on the left of the diagram.
(c) Convince your advisor that your labels are correct by describing the aspects of chromatin structure that are demonstrated by each lane.
Lane 1 : 10mg/ml
Lane 2 : 0mg/ml
Lane 3 : 1mg/ml
Lane 4 : 0.1mg/ml
Marker (bottom to top) = ~50, 200, ~350, ~700
Lane 1 = complete digestion of chromatin with DnaseI showing the 10bp periodicity of cutting of nucleosomes
Lane 2 = uncut chromatin
Lane 3 = near complete digestion of chromatin to mononucleosome sized fragments
Lane 4 = partial digestion with Dnase I showing mono-, di-, tri- and tetranucleosomal fragments