AbBCIC FACS protocol2
advertisement

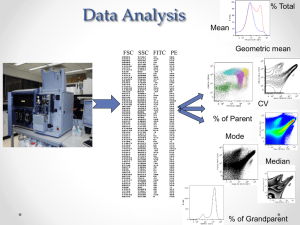
BCIC FACS protocol For antibodies: FITC ESA PE CD24 APC CD44 BD# 347197 lot: 25540 BD# 555428 lot: 08894 BD# 559942 lot: 01798 clone: EBA-1 isotype: Ms IgG1, clone: ML5 isotype: Ms IgG2a, clone: G44-26 isotype: Ms IgG2b, Media: the media, the cells of interest usually live in FACS buffer: PBS with 1 – 3% CS Prepare a single cell suspension from the cell line of interest (in media) Count the cells and resuspend them at 1x106 cells per mL (media) Transfer L cell suspension into FACS tubes (if you feel more comfortable, you could just stain 150 000 cells; 150L) The following stainings are mandatory: Per cell line: Unstained sample Isotype control tube with FITC, PE and APC Single FITC Single PE Single APC BCIC stained sample For subsequent samples: Unstained sample BCIC stained sample Wash with FACS buffer Either decant (leaves 100L cells suspension) or remove the supernatant and resuspend in 100 - 250L FACS buffer Prepare a master mix of antibodies and one with isotype with: 10L ESA FITC 5L CD24 PE 5L CD44 APC per sample (adjust if 150 000 cells are stained) Add the correspondent antibody into the tubes (see above) and 20L antibody mix/isotype mix to the BCIC profile tube Flick, flick, flick, flick Incubate 10min at RT Wash twice with ice cold FACS buffer and resuspend in 250L for analysis At the FACS machine: Prepare your sheet that you can see a FSC/SSC (lin mode) dot plot, a FITC/PE dot plot (to check for auto fluorescence “tail”) and each color as a histogram This just needs to be done once; you can save it on your USB stick for further use Use the setup mode of the machine to look at your unstained sample (autofluorescence control) and position the cells of interest somewhere into the lower/left area of the FSC/SSC dot plot Here some cell line references: sum149 FSC: 430, SSC: 380 sum159 FSC: 405, SSC: 470 BT20 FSC: 350, SSC: 450 MDAMB361 FSC: 350, SSC: 480 HCC70 FSC: 365, SSC: 490 Then check every channel (histogram) and move the population into the area between 100 and 101 (negative population) Check the isotype control tube (unspecific binding control) Make sure that the compensation matrix is set to 0 Now collect 30000 events for all the tubes Analysis with flowjo: import your samples of interest the following steps are done for the compensation: open the isotype control tube and gate for your cell population (you can name it compensation populations) open a dot plot with FSC (x axis) and FITC (y axis) create a rectangle gate on the population including at least 98% of the cells (name it FITC neg) change the y axis to PE and repeat the negative gating (PE neg) drag and drop the “compensation populations” gate from the isotype control into the single stained samples open the single FITC stain and draw a gate on the FITC pos population using a FITC vs. FCS dot plot repeat this steps for PE single stained open → tools → compensation → define new matrix name your matrix drag and drop your populations from the samples into the corresponding box of the matrix (just FITC and PE, because APC does not need to be compensated, if you work with PerCP also the compensation needs to be done for all colors) click “compute and apply” and “yes” for all samples by the mark in front of the samples you can see, that a compensation is set for each sample save your matrix (chose the “edit matrix” tab on the top) set your markers for analysis on compensated samples: Histogram method: open you isotype sample and drag and drop your “compensation populations” gate into any population to generate a copy, rename it to “cells of interest” and drag and drop it back to the parent and delete it from the subpopulation (if anybody figures out how to create a gate copy in an easier way, please let me know...) open the “cells of interest” population in a histogram window with compensated FITC on the x axis create a bifurcated gate such that the negative marker includes at least 98% change the x axis to compensated PE and repeat the gating change the x axis to APC and repeat the gating You now should have a “cells of interest” population with neg and pos populations for each color (something like this...) mark all populations and drag and drop them into your BCIC stained sample drag and drop the CD24- into the CD44+ population and the ESA+ into the just generated CD24- population add statistics for frequency of “cells of interest”