Climatic and evolutionary drivers of community structure in New
advertisement

Appendix S1
Polytomies were generated by directly editing the NEWICK tree. For example, consider a
tree with three genera, A, B, and C, where A and B are sister taxa that diverge 10 time
units before the present, and both diverged from the lineage that gave rise to C 15 time
units before the present. Thus, the tree is described as ((A:10,B:10):5,C:15) (Fig. S1a). If
three representatives of genus A were present in the community, we explored the
consequences of considering each species within A as a basal polytomy (i.e., each species
equally divergent to one another as they are to the sister genus) and a terminal polytomy
(i.e., each species with zero divergence time among them). Thus, the basal polytomy is
described as (((A1:10,A2:10,A3:10):0,B:10):5,C:15) (Fig. S1b), and the terminal
polytomy is described as (((A1:0,A2:0,A3:0):10,B:10):5,C:15) (Fig. S1c). It should be
noted that these polytomies represent the unrealistic extremes of the distribution of all
possible topologies and timing of cladogenetic events.
1
Appendix S2
Original and substitute genera included in the phylogeny. The last column indicates the
number of time each genera had to be substituted for the analyses of phylogenetic
community structure.
No. of
substitutions
Original Genus Substituted
Reference
3
NOT INCLUDED (Fernandez 2004)
Adelomyrmex
19
(Schultz & Brady 2008)
Atta
Acromyrmex
9
(Bolton 2003)
Carebara
Pheidologeton
2
(Bolton 2003)
Carebarella
Monomorium
(De Andrade & Baroni Urbani 6
Cephalotes
Procryptocerus
1999)
54
Cyphomyrmex Trachymyrmex
(Schultz & Brady 2008)
2
Formicoxenus
Leptothorax
(Francoeur et al. 1985)
5
Harpagoxenus Cardiocondyla
(Bolton 2003)
2
Hylomyrma
Pogonomyrmex
(Bolton 2003)
1
Labidus
Eciton
(Brady 2003)
2
Megalomyrmex Monomorium
(Bolton 2003)
1
Mycocepurus
NOT INCLUDED (Schultz & Brady 2008)
10
Nomamyrmex
Eciton
(Brady 2003)
101
Ochetomyrmex Temnothorax
(Bolton 2003)
49
Ponera
Hypoponera
(Yoshimura & Fisher 2007)
1
Protomognathus Temnothorax
(Bolton 2003)
7
Rogeria
Stenamma
(Bolton 2003)
5
Sericomyrmex
Trachymyrmex
(Schultz & Brady 2008)
1
Tranopelta
Pheidologeton
(Bolton 2003)
Brady S.G. (2003) Evolution of the Army Ant Syndrome: The Origin and Long-Term
Evolutionary Stasis of a Complex of Behavioral and Reproductive Adaptations.
Proceedings of the National Academy of Sciences of the United States of America,
100, 6575-6579
2
De Andrade M.L. & Baroni Urbani C. (1999) Diversity and Adaptation in the Ant Genus
Cephalotes, Past and Present. . Stuttgarter Beiträge zur Naturkunde Serie B
(Geologie und Paläontologie), 271, 1-889
Fernandez F. (2004) Adelomyrmecini New Tribe and Cryptomyrmex New Genus of
Myrmicine Ants (Hymenoptera : Formicidae). Sociobiology, 44, 325-335
Francoeur A., Loiselle R. & Buschinger A. (1985) Biosystematique De La Tribu
Leptothoracini (Formicidae, Hymenoptera). 1. Le Genre Formicoxenus Dans La
Region Holarctique. Le Naturaliste Canadien, 112, 343-403
Schultz T.R. & Brady S.G. (2008) Major Evolutionary Transitions in Ant Agriculture.
Proceedings of the National Academy of Sciences of the United States of America,
105, 5435-5440
Yoshimura M. & Fisher B.L. (2007) A Revision of Male Ants of the Malagasy Region
(Hymenoptera : Formicidae): Key to Subfamilies and Treatment of the Genera of
Ponerinae. Zootaxa, 21-40
3
Appendix S3
R scripts used to generate NRI values using four different definitions of the source pool,
implemented in null analyses of phylogenetic community structure.
richness <- function(mat) apply(mat,2,sum)
incidence<- function(mat) apply(mat,1,sum)
species_index <- function(mat,site) return(which(mat[,site]==1))
species_index_allmat <- function(mat)
{
temp<-mat
incidence<-incidence(temp)
return(which(incidence>0))
}
df_sites_index <- function(mat,sp_index)
{
temp<-mat[sp_index,]
rich <- richness(temp)
return(which(rich>0))
}
df_species_index <- function(mat,d_sit_index)
{
temp<-mat[,d_sit_index]
inc <- incidence(temp)
return(which(inc>0))
}
get_df_species_index <- function(mat,site)
{
return(df_species_index(mat,df_sites_index(mat,species_index(mat,site))))
}
df_sample_weights <- function(mat,sp_index)
{
temp<-mat[sp_index,]
return (richness(temp))
}
pick_species_at_site<-function(mat,site)
{
species <-species_index(mat,site)
4
indices <- sample(species,1)
return(indices)
}
null_0s <- function(mat, site)
{
source_pool <-species_index_allmat(mat)
empirical_richness <- sum(mat[,site])
incidence <- incidence(mat)[source_pool]
community_index <- sample(source_pool, empirical_richness, prob = NULL)
return (community_index)
}
null_1 <- function(mat, site)
{
source_pool <- get_df_species_index(mat, site)
empirical_richness <- sum(mat[,site])
community_index <- sample(source_pool, empirical_richness)
return (community_index)
}
null_2w <- function(mat, site)
{
source_pool <- get_df_species_index(mat, site)
df_field_sites <- df_sites_index(mat,species_index(mat,site))
empirical_richness <- sum(mat[,site])
incidence <- incidence(mat[,df_field_sites])[source_pool]
community_index <- sample(source_pool, empirical_richness, prob = incidence)
return (community_index)
}
null_3 <- function(mat, site)
{
species_in_focal_cell <- species_index(mat,site)
sites <- df_sites_index(mat, species_in_focal_cell)
weight <- df_sample_weights(mat, species_in_focal_cell)[sites]
empirical_richness <- sum(mat[,site])
community<-vector()
for (i in 1:empirical_richness)
{
random_species <- 0
while (TRUE)
{
random_site <- sample (sites, 1, prob = weight)
random_species <- pick_species_at_site(mat, random_site)
5
if (sum(!is.na(match(community,random_species))) == 0) ##MKBJUN4: this code
was in error!
break
}
community[i] <- random_species
}
return (community)
}
simulate_phylogenetic_dispersion_vals <- function(SP_FUN, mat, site, phylomat)
{
rand_com <- SP_FUN(mat, site)
mpd <- calculate_mpd(rand_com, phylomat)
mnnd <- calculate_mnnd(rand_com, phylomat)
return(cbind(mpd, mnnd))
}
null_distributions <- function (SP_FUN, mat, site, phylomat, reps = 99)
{
null_dist <- replicate(reps, simulate_phylogenetic_dispersion_vals(SP_FUN, mat, site,
phylomat))
null_dist <- as.matrix(null_dist)
rownames(null_dist) <- c("mpd","mnnd")
return(t(null_dist))
}
show_distributions <- function(SP_FUN, mat, site, phylomat, reps = 100)
{
n <- null_distributions(SP_FUN, mat, site, phylomat, reps-1)
emp_mpd <- calculate_mpd(species_index(mat,site),phylomat)
emp_mnnd <- calculate_mnnd(species_index(mat,site),phylomat)
mpd_dist <- sort(append(n[,1],emp_mpd))
mnnd_dist <- sort(append(n[,1],emp_mnnd))
par(mfrow = c(1,2))
hist(mpd_dist, xlab = "Mean phylogenetic distance", main =
paste(colnames(mat)[site]))
abline(v = emp_mpd, lty = 2)
hist(mnnd_dist, xlab = "Nearest neighbor distance", main = paste(colnames(mat)[site]))
abline(v = emp_mnnd, lty = 2)
return(c(mean(which(mpd_dist == emp_mpd)) / reps, mean(which(mnnd_dist ==
emp_mnnd)) / reps))
}
effect_size <- function(observed, expected, stdev)
{
6
return (-(observed-expected)/stdev) }
rank_value <- function(simulated_values, empirical_value)
{
distribution <- append(simulated_values, empirical_value) ##MKBJUN4: I add the
empirical value to the distribution
distribution <- sort(distribution)
rank <- 0
if (empirical_value == min(distribution)) rank <- 0
else
if (empirical_value == max(distribution)) rank <- 1
else
rank <- mean(which(empirical_value == distribution))/ length(distribution)
return (rank)
}
create_val <- function(mat, phylomat, site, SP_FUN, reps = 100)
{
n <- null_distributions(SP_FUN, mat, site, phylomat, reps-1)
if (richness(com)[site] < 2)
#MKBJUN6: If only one species has been found, it
is not possible to calculate phylogenetic dispersion
return(rbind(NA,NA,NA,NA))
emp_mpd <- calculate_mpd(species_index(mat,site),phylomat)
emp_mnnd <- calculate_mnnd(species_index(mat,site),phylomat)
rank_mpd <- rank_value(n[,1], emp_mpd)
rank_mnnd <- rank_value(n[,2], emp_mnnd)
ef_size_mpd <- effect_size(emp_mpd,mean(n[,1]),sd(n[,1]))
ef_size_mnnd <- effect_size(emp_mnnd,mean(n[,2]),sd(n[,2]))
return(rbind(rank_mpd,ef_size_mpd, rank_mnnd, ef_size_mnnd))
}
7
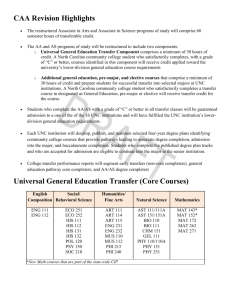
Table S1
Four generalized linear models of phylogenetic community structure (i.e., net relatedness index). Each Generalized Linear Model
includes NRI values generated using one of four definition of the source pool. Stars indicate the level of statistical significance (*
P<0.01,** P<0.001,*** P<0.0001).
full
Estimates
dispersion field
95% CI
0.990
1.896
Estimates
1.033***
95% CI
0.669
1.396
incidence-weighted
estimates
1.282
3.404
estimates
95% CI
Intercept
1.443***
Min.Temp.
-0.006*** -0.008 -0.005 -0.003*** -0.004 -0.002 -0.003*** -0.006 -0.003 -0.002* -0.003 -0.001
-0.001 -4.40 e-5
-0.001 -0.000
Min.Tem × Precip.
2.56e-6*
5.99e-7 4.53e-6 2.50 e-6* 9.29 e-7 4.08 e-6 2.47 e-6* 7.76 e-7 4.17 e-6 1.85 e-6* 4.83 e-7 3.21e-6
0.13
0.12
-0.000
0.854
-0.001*
0.23
-0.001** -0.001 -0.000
0.540** 0.226
Precip.
r2 observed~predicted
-0.001** -0.001 -0.000
1.002***
95% CI
similarity-weighted
0.06
Table S2
Four generalized linear models of ant taxonomic richness. Each Generalized Linear Model includes taxonomic richness values
at the level of the species, genus, tribe or subfamily. All models are with a Poisson distribution and a log link function. Stars
indicate the level of statistical significance (* P<0.01,** P<0.001,*** P<0.0001).
species
Estimates
Intercept
Min.Temp.
Precip.
Min.Tem × Precip.
r2 observed~predicted
2.90***
genus
95% CI
0.990
1.896
Estimates
tribe
95% CI
estimates
subfamily
95% CI
estimates
95% CI
2.366***
2.257
2.474 2.085*** 1.964 2.203 1.219*** 1.036 0.396
0.002*** -0.008 -0.005 0.003***
0.002
0.003 0.002*** 0.001 0.002 0.002*** 0.001 0.003
-0.000*** -0.000 -0.000 -0.000*** -0.000 -0.000
n.s.
n.s.
n.s
n.s.
n.s.
n.s
1.31e-6*** 9.17e-7 1.69e-6 5.79 e-6 5.79 e-8 1.02 e-6
n.s.
n.s.
n.s
n.s.
n.s.
n.s
0.16
0.30
0.18
0.31
Figure S1.
Scenario for generated polytomies when multiple species within a genus are present in a community. a) Phylogeny with three
genera. b) Phylogeny with three genera and three representatives of genus A forming a basal polytomy. c) Phylogeny with
three genera and three representatives of genus A forming a terminal polytomy.
Figure S2.
This figure shows the distribution of the frequency of species incidence across all communities. Incidence is the number of
communities in which a species was recorded and frequency indicates the number of times a species fell in one of the incidence
categories.
.
Figure S3.
Taxonomic richness is, generally, negatively related to the degree of phylogenetic
clustering among North American ant communities. The strength of the relationship
between ant richness ant phylogenetic structure increases with decreasing taxonomic
resolution.
12
Figure S4.
The phylogenetic signal in the climatic niches of North American ants. The phylogenetic
tree includes 591 species. Color labels at the tip of each branch indicate (A) the minimum
temperature of the coldest month and (B) the minimum annual precipitation recorded for
a species across all communities at which it occurred. Three temperature and
precipitation categories were created to facilitate the visualization of climatic extremes on
the phylogenetic tree.
A
B
13