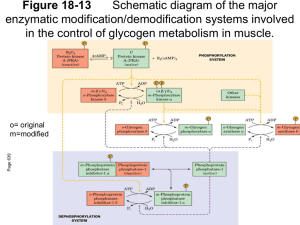
Drug Discovery and Drug Development in the
advertisement

Drug Discovery in the Kinase Inhibitory Field using the Nested Chemical Library TM technology György Kéria,b*, Zsolt Székelyhidia, Péter Bánhegyia, Zoltán Vargaa, Bálint-Hegymegi Barakonyia, Csaba Szántai-Kisa, Doris Hafenbradld#, Bert Klebld#, Gerhard Mullerd#, Axel Ullriche, Dániel Erősb, Zoltán Horváthb, Zoltán Greffb, Jenő Marosfalvib, János Patób, István Szabadkaib, Ildikó Szilágyib, Zsolt Szegedib, István Vargab, Frigyes Wáczeka, László Őrfia,b,c a Peptide Biochemistry Res. Group of Hung. Acad. Sci. and Cooperative Research Centre, Semmelweis University, Rippl-Rónai u. 37., Budapest 1062, Hungary b c Vichem Chemie Research Ltd., Herman Otto u. 15., Budapest 1022, Hungary Department of Pharmaceutical Chemistry, Semmelweis University, Hőgyes Endre u. 9., Budapest 1092, Hungary d # Axxima Pharmaceuticals AG, Max-Lebsche-Platz 32, Munich 81377, Germany now GPC Biotech AG, Max-Lebsche-Platz 32, D-81377 Munich, Germany e Max Planck Institute, Department of Biochemistry, D-82152, Am Klopferspitz 18/A, Martinsried , Germany * Corresponding author: Tel.:+36-1-487-2080; fax: +36-1-487-2081; e-mail: keri@vichem.hu Abstract Kinase inhibitors are in the front line of modern drug research where mostly three technologies are used for hit and lead finding: HTS of random libraries, 3D design based on X-ray data, and focused libraries around limited number of new cores. Our novel Nested Chemical Library (NCL) technology is based on a knowledge base approach where focused libraries around 1 selected cores are used to generate a pharmacophore model. NCL was designed on the platform of a diverse kinase inhibitory library organized around 97 core structures. We have established a unique proprietary kinase inhibitory chemistry around these core structures with small focused sublibraries around each core. All of the compounds in our NCL library are stored in a big unified SQL (Structured Query Language) database along with their measured and calculated physicochemical and ADME and toxicity (ADMET) properties, together with thousands of molecular descriptors calculated for each compound. Drug-likeness of all the compounds can be visualized with the widely accepted calculated Lipinski parameters. Biochemical kinase inhibitory on selected cloned kinase enzymes for a few hundred compound sets from NCL can provide enough biological data for rational computerized design of new analogues based on our pharmacophore model generating 3DNET4WTM QSPAR (Quantitative Structure- Property/Activity Relationships) approach. Using this pharmacophore modelling approach and the ADMET filters we can preselect the synthesizable compounds for hit and lead optimisation. Starting from there and integrating the information from QSPAR high quality leads can be generated within a small number of optimization cycles. Introduction In recent years, dramatic developments in molecular biology and protein chemistry have greatly enhanced our understanding of the molecular pathomechanisms of various diseases. As a result, traditional concepts of drug design that apply trial-and-error approaches are being increasingly superseded by novel strategies. These are based on the knowledge of molecular pathomechanisms related to the disorders of inter- and intracellular signalling.1 More than 80 2 percent of diseases with known molecular pathomechanisms involve communication abnormalities, including cancer, infectious diseases, arteriosclerosis, arthritis, neurodegenerative disorders, etc. False proliferation signals, occupied signaling channels, cell-injury, inflammation, autoimmune reactions are potential factors in the etiology of these diseases.2 Signal transduction therapy endeavours to repair signaling defects involved in cell communication. A communication system works properly if the shared information is relevant and the individual components of the system contribute adequate responses. Communication is prerequisite to the maintenance and development of living systems and any malfunction of intra and intercellular communication leads to disease. Normal cells that duly perform their physiological functions do not send or receive false messages and are securely controlled by the messages of the communication network. They are even willing to “commit suicide” (apoptosis) for the sake of the host organism. This situation is entirely different with diseased cells. Cancer cells, for example, generate a false, mimicked proliferation signal for themselves (via oncogenes and other genomic changes) and these results in their uncontrolled growth, while the so called survival factors inhibit the programmed cell death mechanism.3,4 In recent years many validated target molecules have been discovered in various pathological states especially in cancer and many more are expected in the near future. The human genome is reported to contain about 32,000 genes that express an estimated 250,000 to 300,000 proteins. The increased number of proteins might be explained by through alternative splicing and post transcriptional and translational modifications and regulations. About 20-25 percent of the druggable genome are kinases involved in signal transduction, but at this stage only 4 kinase inhibitors are in clinical practice, which means that the field still offers a lot of options for drug discovery. One of the first proof of concept drugs for signal transduction therapy in cancer was 3 Gleevec – an inhibitor of Bcr-Abl kinase – launched in May 2002.5, 6 Gleevec treats chronic myeloid leukemia (CML) with 90-per-cent efficacy. According to our present knowledge almost 200 compounds with kinase inhibitory activity are in preclinical and clinical development against more than 50 kinase targets for signal transduction therapy (the current clinical pipeline contains 53 small molecule kinase inhibitors)7 this definitely indicates the significance influence of this area in drug research.8, 9 The most important small molecule synthetic tyrosine kinase inhibitors which have been launched or are in late stage clinical development for cancers are Gleevec, the epidermal growth factor receptor tyrosine kinase inhibitors: Iressa, Tarceva and Lapatinib, the BRaf inhibitor BAY-43-9006 and the VEGF kinase inhibitor, SU11248. Several agents targeting other tyrosine kinases implicated in cancer, are in preclinical development.10 Our team contributed significantly to the development of SU101 which has reached Phase III. Clinical trials and to the predecessor oxindols of SU11248, one of the most promising clinical candidates in cancer trials, which addresses inhibition of the tyrosine kinases PDGF-R, Flt-3, VEGF-R, and c-kit11, 12, 13 Hit and lead finding strategies against validated targets can be achieved via high throughput screening of huge combinatorial chemistry libraries or via rational drug discovery. Theoretically combinatorial chemistry can result in extreme numbers (10100) of planned compounds. This number can be reduced by applying pertinent restrictions (i.e. exclusion of larger molecules and using Lipinski’s rules); however, approximately 1050 molecules remain after applying such filters, which means that combinatorial chemistry still means as much as shooting in the dark. This confirms the conclusion that testing random libraries for hit and lead finding cannot be efficient and rational drug design is thus the preferred scenario for kinase inhibitor drug discovery.14 The scheme of rational drug design is illustrated in Figure 1. 4 Figure 1. Scheme of rational drug design (HTS: High Throughput Screening, RDD: Rational Drug Design). Originally, the term rational drug design was used for the planning of molecules, based on the three-dimensional structure of targets. In the meantime, however, the concept of rational drug design has been expanded and now it means the whole complex process including pathomechanism-based target selection, target validation, structural biology, molecular modelling, structure-activity relationships, pharmacophore-based compound selection and pharmacological optimisation.15 Materials and Methods Vichem Chemie Research Ltd has developed a unique hit and lead finding method based on its Nested Chemical LibraryTM (NCL) technology (Figure 2.) The NCL was designed on the platform of our up-to-date knowledge base what we have built up from the knowledge we 5 accumulated from our experience in the last 15 years of kinase inhibitory chemistry. Our library is organized around 97 core structures and we have generated small focused sublibraries around each core. CVL: Chemical Validation Library EVL: Extended Validation Library ML: Master Library Figure 2. Nested Chemical Library™ (NCL) The Chemical Validation Library (CVL) includes ~300 compounds around 97 selected core structures with proven kinase inhibitory activity on various kinase targets arising from work published in the current literature and patent(s) (applications). (Figure 3 shows 10 representative core structures) 6 N N N N 1. 6. 36. N N 16. N 29 N N N N N O 13. N N N N N N 37. 38. O 54. 86. Figure 3. NCL is clustered around 97 core structures, 10 representative structures are shown EVL is an extension of the CVL and covers almost 700 compounds based on the following criteria: to increase the number of target kinases against which we have inhibitors available (currently we synthesized inhibitors for 83 different kinases) and to include more analogues around the proven kinase inhibitory leads from the CVL. The Master Library (ML) includes novel compounds and analogue structures around CVL and EVL, extended by an additional ~ 10 000 compounds. We have a large series of new and patentable compounds in ML with a lot of space for further chemical and pharmacological optimisation. Typically, novel and potential kinase targets can be validated chemically by using the Chemical Validation Library compounds16, 17, 18 . CVL compounds are tested in biochemical assay for activity against a kinase of interest. Next, the selected and active inhibitors of a particular kinase are applied to therapeutically relevant cellular assays in order to confirm the role of the kinase activity in this biological model. The validated hits from the CVL will also represent a good starting point for extended biological screening. Biochemical activity data obtained for the selected compound sets from NCL can provide enough biological data for 7 rational computerized design of new analogues based on our Pharmacophore model generating 3DNET4WTM QSPAR approach.19,20,21 The program builds abstract pharmacophore (QSAR) models from validated, significant descriptors selected by sequential or genetic algorithms. Enhanced MLR (Multiple Linear Regression), PLS (Partial Least Squares), ANN (Artificial Neural Network), LLM (Linear Learning Machine) methods are optional and can be combined with PCA (Principal Component Analysis). Pharmacophore models are validated by external validation, the validation set is not known for the program. New analogues are designed on the basis of bioisosteric changes using validated standard reaction schemes. New potential hit molecules are filtered by their ADMET properties, drug-likeness and patentability. Thousands of molecular descriptors are calculated for hit compound structures and fed with 3DNT4WTM QSPAR program system. In the hit and lead optimisation work interesting compounds are selected by our pharmacophore model and the ADMET filters. This in silico selection subsequently becomes synthesized. The new compounds are then tested for biological activity, physicochemical parameters are being calculated and measured . These data are fed back into the model for continuous improvement and optimization. If we have good and reliable biochemical kinase assays available, we can generate a lead compound against a particular target molecule within approximately five chemical optimization cycles. For optimisation of the hit and lead molecules we apply two methods: focused and diverse optimisation. If the hit compound is patentable we apply focused optimisation: generating various possible derivatives around the selected leads with traditional medicinal chemistry approach. In case of the hit compound is not patentable we generate diverse molecular library with virtual screening, pre-selecting them with the pharmacophore model. The new compounds 8 are synthesised with solution or solid-phase synthesis, validated with HPLC-MS and NMR and tested in biological assay. The biological results are again fed back into the pharmacophore model to accomplish the lead optimisation process. In summary, approximately five compound optimization cycles are needed with this approach to generate a series of sufficiently characterized lead molecules against a kinase target. (Figure 4.). 22, 23, 24, 25, 26 Figure 4. Hit / Lead Finding Strategy In Table 1 we present a series of validated kinase target molecules and the IC50 values of our best hit compounds (without disclosing the structure but indicating the core structure). In table 2 we present the hit rate, which we found using a series of compounds from our CVL and EVL library against various kinases. 9 Table 1. Targets and inhibitors according to Core structures Number of Target Indica-tions tested Hit compounds Core AX3359 IC50 = 25 pM 13. AX12257 IC50 = 8 pM 58. AX13237 IC50 = 3 nM 16. AX9041 IC50 =10 nM 4. AX13234 IC50 = 20 nM 4. AX13985 IC50 = 77 nM 89. AX11466 IC50 = 13 nM 32. compounds EGF-R PDGF-R Cancer Cancer, 1796 580 gliomas VEGF-R RIP(RIPK-1) Oncology Inflammation 340 724 related diseases RICK Viral (RIPK-2) infections (HCMV), AX11443 IC50 = 16 nM 664 AX8652 IC50 = 40 nM AX7015 IC50 = 40 nM Inflammation related diseases 10 29. UL-97 Viral 927 AX11922 IC50 = 0.4 nM 4. AX7100 IC50 = 13 nM 25. AX39010 IC50 = 7 nM 85.20,27 infections (HCMV) PknG SRPK-1 M. 627 Tuberculosis AX38833 IC50 = 15 nM infection AX38824 IC50 = 15 nM Oncology 432 AX2926 IC50 = 8 nM 16. AX8757 IC50 = 43 nM Akt Oncology 349 AX971 IC50 = 33 nM 8. AX9385 IC50 = 1.1 nM ROCK-2 Oncology, 325 neural AX6115 IC50 = 0.2 mM 1. AX7425 IC50 = 0.7 mM inflammation and regeneration Table 2. Number of tested compounds on kinase panel Target Tested Active Target Tested Active Abl 77 10 PDGFR 54 2 11 Akt 547 37 RICKK 664 64 C-Met 22 2 PKCa 195 14 CDK-1 249 14 PKnG 7435 165 CLK-1 206 41 Rip 724 146 CLK -2 209 19 ROCK-2 325 23 CLK -3 209 6 Src 75 4 InsR 250 9 SRPK-1 432 137 p38 64 7 SRPK-2 582 77 P70 S6K 209 14 UL97 927 442 Results and Discussion Signal transduction therapy is in the front line of modern drug research and kinases represent these days the most important target molecule family. More than 50 kinases have been identified as therapeutically relevant target molecules in various pathological cases including cancer, atherosclerosis, infectious diseases, neurodegenerative disorders and inflammatory diseases. Because cellular signaling occurs via network signaling in several cases a multiple target approach has to be considered. In cancer a series of kinases act like survival factors thus providing primary targets for drug research. There is a good chance to treat various cancers with potent compounds against the most important oncogenic signaling pathways . Furthermore molecular diagnostic techniques serve to analyse the oncogenic pathways in the particular patient in the a so-called personalized therapy where each patient finally would receive a cocktail of the relevant inhibitors for his disease. Since kinases seem to have a major role in oncogenic 12 signaling the perspective is that we should have good approved drugs for the most important oncogenic kinases. As a general strategy for hit and lead finding against a novel kinase target we prefer to screen focused libraries or to determine the 3D structures of the target kinase and perform docking experiments. Our approach is to utilize our hit finding library and using these assay data and calculating large series molecular descriptors for QSAR and pharmacophore model generation. During our hit and lead optimisation process we utilize (and continuously improve) this pharmacophore for pre-selection of the synthesizable compounds. We also use various ADME filtering models we generated based on experimentally determined parameters (from literature and from our own resources) to pre-select the synthesizable compounds not only for activity but also for drug likeliness predictions. Thus we can significantly reduce the number of to be synthesized compounds for a lead optimisation process and can come up with a promising lead candidate within approximately 5 optimization cycles. Acknowledgements References 1. 2. 3. Hanash S: Disease proteomics. Nature 2003; 422: 226-232. Levitzki A: Signal transduction therapy- a novel-approach to disease management. European Journal of Biochemistry 1994; 226: 1-13. Hollósy, Ferenc; Mészáros, György; Bökönyi, Gyöngyi; Idei, Miklós; Seprődi, Attila; Szende, Béla; Kéri, György: Cytostatic, cytotoxic and protein tyrosine kinase inhibitory activity of ursolic acid in A431 human tumor cells. Anticancer Research 2000; 20(6B), 4563-4570 13 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. Kéri Gy.; Rácz G.; Magyar K.; Örfi L.; Horváth A.; Schwáb R.; Hegymegi B B; Szende B: Pro-apoptotic and anti-apoptotic molecules affecting pathways of signal transduction. Annals of the New York Academy of Sciences (2003 Dec), 1010 109-12. Deininger M.W., Goldman J.M., Lydon N., Melo J.V: The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL-positive cells. Blood 1997; 90: 3691-3698. Goldman J.M: Tyrosine-kinase inhibition in treatment of chronic myeloid leukaemia. Lancet 2000; 355: 1031-1032. Klebl B., G. Müller, 2005 Experimental Opinion on Therapeutic Targets (manuscript submitted) Dancey, J., Sausville, E. A: Issues and progress with protein kinase inhibitors for cancer treatment. Nature 2003; 2: 296-313. Kolibaba K.S., Druker B.J: Protein tyrosine kinases and cancer. Biochim. Biophys. Acta 1997; 1333: F217-F248. Laird AD, Cherrington JM: Small molecule tyrosine kinase inhibitors: clinical development of anticancer agents.Expert Opin Investig Drugs. 2003 Jan;12(1):51-64. Strawn L.M., McMahon G., App H., Schreck R., Kuchler W.R., Longhi M.P., Hui T.H., Tang C., Levitzki A., Gazit A., Chen I., Kéri Gy., Őrfi L., Risau W., Flamme I., Ullrich A., Hirth K.P., Shawver L.K: Flk-1 as a Target for Tumor Growth Inhibition. Cancer Research 1996; 56: 3540-3545. Hirth, K.P., Shawver, L.K., Kéri, Gy., Örfi, L., Székely, I., Haimichael, J., Ullrich, A., Lammers, R: Treatment of platelet derived growth factor related disorders such as cancers U.S. Patent No.: 5990141 Morimoto AM, Tan N, West K, McArthur G, Toner GC, Manning WC, Smolich BD, Cherrington JM Gene expression profiling of human colon xenograft tumors following treatment with SU11248, a multitargeted tyrosine kinase inhibitor. Oncogene. 2004 Feb 26;23(8):1618-26 Beroza P., Villar H.O., Wick M.M., Martin G.R: Chemoproteomics as a basis for post genomic drug discovery. Drug Discov. Today 2002; 7: 807-814. Dániel Erős, László Őrfi, György Kéri: Rational Drug design and signal transduction therapy. Pharmachem 2004; 1. Őrfi, László; Wáczek, Frigyes; Kövesdi, István; Mészáros, György; Idei, Miklós; Horváth, Anikó; Hollósy, Ferenc; Mak, Marianna; Szegedi, Zsolt; Szende, Béla; Kéri, György: New antitumor leads from a peptidomimetic library. Letters in Peptide Science 1999; 6(5-6), 325-333. Klebl, B. M.: Chemical Kinomics: A Target Gene Family Approach in Chemical Biology. Drug Discovery Today:Technologies. 2004; 1: 25-34. Klebl, B. M.; Daub, H.; Kéri, Gy.: Chemical Kinomics. In: Chemogenomics in Drug Discovery: A Medicinal Chemistry Perspective. Kubinyi H, Müller G (Eds.) WILEYVCH Verlag GmbH & Co. KgaA, Weinheim. 2004; 167-190. Erős, D.; Kövesdi, I.; Őrfi, L.; Takács-Novák, K.; Acsády, Gy.; Kéri, Gy: Curr. Med. Chem., 2002; 9, 1819-1829. Erős, D.; Kéri, Gy.; Kövesdi, I.; Szántai-Kis, Cs.; Mészáros, Gy.; Őrfi, L: Mini-Reviews in Medicinal Chemistry, 2004; 4, 167-177. Kövesdi, I.; Kéri, Gy.; Ôrfi, L. WO Patent 02/082329. 14 22. 23. 24. 25. 26. 27. Koul, Anil; Klebl, Bert; Mueller, Gerhard; Missio, Andrea; Schwab, Wilfried; Hafenbradl, Doris; Neumann, Lars; Sommer, Marc-Nicola; Mueller, Stefan; Hoppe, Edmund; Freisleben, Achim; Backes, Alexander; Hartung, Christian; Felber, Beatrice; Zech, Birgit; Engkvist, Ola; Kéri, György; Őrfi, László; Bánhegyi, Péter; Greff, Zoltán; Horváth, Zoltán; Varga, Zoltán; Markó, Péter; Pató, János; Szabadkai, István; Székelyhidi, Zsolt; Wáczek, Frigyes; WO 2005023818 A2 Blencke, Stephanie; Zech, Birgit; Engkvist, Ola; Greff, Zoltán; Őrfi, László; Horváth, Zoltán; Kéri, György; Ullrich, Axel; Daub, Henrik. Axxima Pharmaceuticals AG, Munchen, Germany. Chemistry & Biology 2004; 11(5), 691-701. Godl Klaus, Missio Andrea, Daub Henrik, Stein-Gerlach Matthias, Greff Zoltan. WO 2004013633 Pató, J.; Kéri, Gy.; Örfi, L.; Wáczek, F.; Horváth, Z.; Bánhegyi, P.; Szabadkai, I.; Marosfalvi, J.; Hegymegi-Barakonyi, B.; Székelyhidi, Zs.; Greff, Z.; Choidas, A.; Bacher, G.; Daub, H.; Obert, S.; Kurtenbach, A.; Habenberger, P. WO Patent 02/094796 A2, 2002. Örfi, László; Wáczek, Frigyes; Pató, János; Varga, István; Hegymegi-Barakonyi, Bálint; Houghten, Richard A.; Kéri, György: Improved, high yield synthesis of 3H-quinazolin-4ones, the key intermediates of recently developed drugs. Current Medicinal Chemistry (2004; 11(19), 2549-2553. János Pató, György Kéri, László Örfi, Frigyes Wáczek, Zoltán Horváth, Péter Bánhegyi, István Szabadkai, Jenő Marosfalvi, Bálint Hegymegi-Barakonyi, Zsolt Székelyhidi, Zoltán Greff, Axel Choidas, Gerald Bacher, Andrea Missio, Anil Koul; US 2004/0171603 A1 15