Appendix S1
advertisement

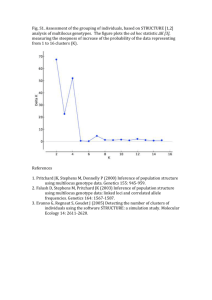
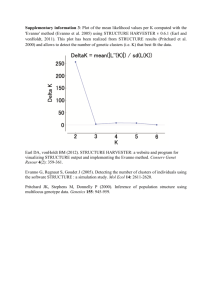
Appendix S1 – 1 Appendix S1. Genetic structure of Helleborus foetidus plants sampled for the study Methods To examine whether the 60 H. foetidus plants sampled for this study from three locations (TEJ, NAV, and PLL) were genetically differentiated, and to define the most likely number of genetic clusters (K) represented in the sample, we used Bayesian clustering analyses as implemented in STRUCTURE v2.3 (Pritchard et al. 2000; Falush et al. 2003, 2007). These analyses were performed on the data set of 103 fragments obtained with the conventional AFLP fingerprinting technique, using the following parameters: length of burn-in period = 50,000, number of Markov chain Monte Carlo reps after burn-in = 100,000, admixture model, and correlated allele frequencies. To test the stability of the results, 20 iterations per K level were performed from K = 1 through K = 7. The most probable number of genetic clusters was determined by calculating the ad hoc statistic delta K, which identifies the highest rate of change in the log-likelihood between successive Ks (Evanno et al. 2005), in STRUCTURE HARVESTER (Earl & vonHoldt 2012). The program CLUMPP (Jakobsson & Rosenberg 2007) was used to compile individual assignments across all 20 replicates for the most likely K, with the Greedy algorithm and 5000 reps. Individual group membership probabilities for individual plants were plotted for visualization in DISTRUCT 1.1 (Rosenberg 2004). Results The results of the STRUCTURE analyses revealed that the recognition of two clusters (K = 2) best reflected the genetic structure present in our data (Fig. S1A). At K = 2, the two populations located at high and mid elevation (PLL and NAV, respectively) were clearly differentiated from the lowest population (TEJ; Fig. S1B). In the two extreme populations (TEJ and PLL) the vast majority of individuals were assigned to one of these two genetic clusters with a probability of 0.9 or more. Slightly more admixture between these two lineages occurred at the intermediate population (NAV), as indicated by the fact that 45% of the plants had assignment probabilities between 0.69 and 0.89 (Fig. S1B). Appendix S1 – 2 Figure S1. Results of the Bayesian clustering analysis using STRUCTURE of the 60 plants x 103 AFLP loci data matrix. (A) Estimation of the most probable number of genetic clusters present in the sample (K) for K=1-6 and 20 independent STRUCTURE simulations: posterior probabilities of the data ln[P(D)] (open circles) and values of Evanno’s delta K (filled circles). (B) Barplot showing the probability of membership to the two inferred clusters for each of the 60 Helleborus foetidus individuals sampled (vertical lines). Colors represent the estimated probabilities of membership to each cluster. TEJ, NAV and PLL denote the three sampling locations. A B -4000 20 -4400 15 -4800 10 -5200 5 -5600 0 -6000 1 2 3 4 5 Mean LnP(D) Delta K 25 6 K References Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611–2620. Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes, 7, 574–578. Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23, 1801–1806. Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics, 155, 945–959 Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Molecular Ecology Notes, 4, 137–138.