The structure of RNase E at the core of the RNA
advertisement

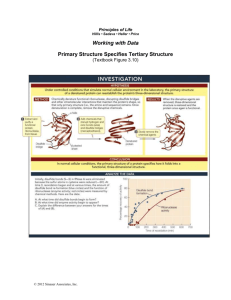
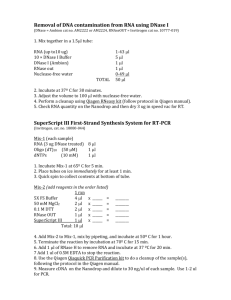
SUPPLEMENTARY INFORMATION Additional commentary on features of the structure As described in the main text, the N-terminal half of E. coli RNase E encompasses the catalytic function of the enzyme1-3, and this portion resembles closely the paralogue, RNase G4,5. The C-terminal half of E. coli RNase E is largely unstructured and poorly conserved6. In E. coli and related proteobacteria, this Cterminal domain organizes and coordinates the activities of the multi-enzyme RNA degradosome complex7,8, which is composed of enolase, polynucleotide phosphorylase and the ATP-dependent helicase RhlB. The degradosome assembly is required for the targeted degradation of transcripts9,10 and the coupled degradation of small regulatory RNAs with their cognate transcript11. Guided by the first structure for the RNase E/G family, a sequence alignment was prepared for representative members of the group. The intricate details of the structure account for the conservation of key residues at the catalytic site, the hydrophobic core, and protein-protein interfaces, as indicated in Fig. S1. A single protomer of RNase E catalytic domain bound to RNA occupies the crystallographic asymmetric unit, and the tetramer is generated through crystal symmetry. An oligomer of four identical subunits is consistent with our observations from solution studies12. The tetramer is organized as a dimer-of-dimers with D2 symmetry. Shared zinc ions organize the two principal dimers. The location and identification of the metal in the crystal structure was corroborated by its signal from the anomalous scattering of X-ray (see Materials and Methods below). The presence of thiol-linked zinc in the RNase E catalytic domain is in agreement with our solution 1 studies using X-ray spectroscopy13. In agreement with our mutagenic studies, the zinc is coordinated at the intra-domain linker through a pair of cysteine residues in the conserved CPxCxGxG motif that occurs throughout the RNase E/RNase G family13 (Fig. S1). A similar motif is found in the metal coordinating sites of the bacterial chaperone DnaJ (PDB entry 1EXK), where a single zinc ion is coordinated within the same subunit, rather than between two different subunits as in the case of RNase E. The protomer can be dissected into a smaller and larger subdomain (Fig. S2a) The principal dimer is formed mostly by the self-complementary interaction of the larger domain of the protomer. The small domain also contributes to the principal dimer through inter-subunit contact with the large domain. Another surface of the small domain forms a self-complementary interface in the association of the two principal dimers to create the tetramer. The larger subdomain can be further subdivided structurally. Fig. S2 shows the isolated subdomains of the RNase E catalytic domain. The RNase H fold is discontinuous with regard to the linear sequence (residues 1-35 and 216-279) and is interrupted by the insertion of two additional folds. The first embedded fold is an S1 domain, which is a widely occurring RNA-binding structural motif. The S1 subdomain of RNase E makes important contacts with the RNA near the cleavage site. In comparison with the isolated S1 domain reported earlier14, there are small adjustments in the positions of sidechains and exposed loops that can be attributed to contacts to the RNA and other subdomains of RNase E. The DNase I domain contains the catalytic site. Through self-association, this domain contributes the greatest hydrophobic interface of the RNase E tetramer, and its dimer mimics DNase I. Aside from its self-association, the small domain (residues 415 to 529) appears to 2 be structurally indivisible. The fold of this small domain is similar to that of a domain of myo-inositol 1-phosphate synthase (1jki chain A, residues 323-422). Mutations of the active site and 5’ sensing pocket The RNA is not processed in the co-crystals studied here, probably because the 2’-O-methyl substituent sterically occludes the strained transition state. The crystal structure was used to identify residues of the catalytic site whose importance is indicated by site direction mutagenesis (summarized in Table S1). These are described in the main text. Phe67, Phe57 and Lys112 of the S1 domain form a basesequestering pocket, and substitution of any of these with Ala was found to abolish catalytic activity of RNase E in vivo15. These observations corroborate the in vitro observations of Diwa and Belasco16. An analysis of mutations in the 5’-sensing pocket will be reported elsewhere (Stead et al.,) The snug fit of the 5’ end into the pocket explains why RNA is protected from cleavage by RNase E if this segment has a tri-phosphate cap or is double-stranded: both forms are sterically excluded, as anticipated by Mackie17. The sensing pocket differs from that of the PIWI domain used in RNA silencing mechanisms18,19. Cognate and non-cognate complexes RNase E preferentially cleaves A+U-rich segments20-22 and it cuts the RNA Ibased substrate, 5’ACAGU*AUUUG, at the site indicated by the asterisk23. This preferred cleavage site interacts with the catalytic magnesium in the complex of the 13-mer with RNase E (5’-pUUUACAGU*AUUUG). The separation between the 5’ sensor and the catalytic site suggests an explanation for the finding that RNase E does 3 not efficiently cleave bonds that are less than 8 nucleotides from the 5’monophosphated end of polyA or polyU substrates24,25. The 10-mer structure also has the same optimal spacing between the 5’ sensing pocket and the catalytic magnesium. As a consequence, the catalytic site of the 10-mer complex faces the sequence 5’pACAGUAUU#UG at the site marked by a hash sign (Fig. S3). The 15-mer spans between the 5’ sensing pocket in a protomer of one tetramer to the catalytic site in the protomer of in a symmetry related tetramer. It’s catalytic site faces the sequence 5’pUUUACAGUAUU#UGUU at the site marked by a hash sign. The protein-RNA interactions for the 15-mer RNA nonetheless are similar to those seen for the 10-mer and 13-mer. The 10-mer and 15-mer structures may represent non-cognate cleavage complexes. In all cases – both preferred and non-cognate complexes - the RNA follows the same stacking arrangement over the surface of the RNA-binding channel, and only one of the bases forms a hydrogen bond with the protein; Lys106 contacts the exocyclic oxygen of the base at position -1 with regard to the scissile phosphate. This direct contact may explain the preference for G or U at this position25. Aside from that contact, there is no sequence recognition per se. We propose that the preference of RNase E for A+U-rich substrates appears to arise mainly from “indirect readout”, through the recognition of the RNA conformation. Thus, substrate preference is due to the propensities of the bases to stack on the 5’ side of the scissile phosphate and then pack at right angles immediately following the cleavage site, which is accommodated more readily for A+U rich sequences. Supplementary Information References 4 1 McDowall, K.J. & Cohen, S.N. The N-terminal domain of the rne gene product has RNase E activity and is non-overlapping with the arginine-rich RNA binding site. J. Mol. Biol. 255, 349-355 (1996). 2 Kido, M. et al. RNase E polypeptides lacking a carboxyl-terminal half suppress a mukB mutation in Escherichia coli. J. Bacteriol. 178, 3917-25 (1996). 3 McDowall K.J., Hernandez R.G., Lin-Chao S. & Cohen SN. The ams-1 and rne3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J. Bacteriol. 175, 4245-4249 (1993). 4 Li, Z., Pandit, S. & Deutscher, M. P. RNase G (CafA protein) and RNase E are both required for the 5 maturation of 16S ribosomal RNA. EMBO J. 18, 2878–2885 (1999). 5 Wachi, M., Umitsuki, G., Shimizu, M., Takada, A. & Nagai, K. Escherichia coli cafA gene encodes a novel RNase, designated as RNase G, involved in processing of the 5' end of 16S rRNA. Biochem Biophys Res Commun. 259, 483-488 (1999). 6 Callaghan, A.J. et al. Studies of the RNA degradosome-organising domain of the Escherichia coli RNase E. J. Mol. Biol. 340, 965-979 (2004). 7 Carpousis, A.J. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans 30, 150-155 (2002). 8 Py, B, Higgins, C.F., Krisch, H.M. & Carpousis, A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381, 169-172 (1996). 9 Bernstein, J. A., Lin, P-H., Cohen, S.N., & Lin-Chao, S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc. Nat. Acad. Sci USA 101, 2758–2763 (2004). 5 10 Morita, T., Kawamoto, H., Mizota, T., Inada, T. & Aiba, H. Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol. Microbiol. 54, 10631075 (2004). 11 Massé, E., Escorcia, F.E. & Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374-2383 (2003). 12 Callaghan, A.J. et al. Quaternary structure and catalytic activity of the Escherichia coli ribonuclease E amino-terminal catalytic domain. Biochemistry 42, 13848-13855 (2003). 13 Callaghan, A.J. et al. The ‘Zn-link’: A metal-sharing interface that organizes the quaternary structure and catalytic site of the endoribonuclease, RNase E. Biochemistry 44, 4667-75 (2005). 14 Schubert, M. et al. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J. Mol. Biol. 341, 37-54 (2004). 15 Diwa A.A., Jiang X., Schapira M. & Belasco J.G. Two distinct regions on the surface of an RNA-binding domain are crucial for RNase E function. Mol. Microbiol. 46, 959-969 (2002). 16 Diwa, A.A. & Belasco, J.G. Critical features of a conserved RNA stem-loop important for feedback regulation of RNase E synthesis. J. Biol. Chem. 277, 20415-20422 (2002). 17 Mackie, G.A. Ribonuclease E is a 5’-end-dependent endonuclease. Nature 395, 720-723 (1998). 6 18. Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666-70 (2005). 19. Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663-6 (2005). 20. McDowall, K.J., Lin-Chao, S. & Cohen, S.N. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 269, 10790-10796 (1994). 21. Lin-Chao, S., Wong, T.T., McDowall, K.J. & Cohen, S.N. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 269, 10797-10803 (1994). 22 Cohen, S.N., & McDowall, K.J. RNase E: still a wonderfully mysterious enzyme. Mol. Microbiol. 23, 1099–1106 (1997). 23 McDowall, K.J., Kaberdin, V.R., Wu, S.W., Cohen, S.N. & Lin-Chao, S. Sitespecific RNase E cleavage of oligonucleotides and inhibition by stem-loops. Nature 374, 287-290 (1995). 24 Walsh, A.P. et al. Cleavage of poly(A) tails on the 3’-end of RNA by ribonuclease E of Escherichia coil. Nucleic Acids Res. 29, 1864-1871 (2001). 25 Kaberdin, V.R. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide-based assay. Nucleic Acids Res. 31, 4710-4716 (2003). 26. Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 46734680 (1994). 7 27. Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 15, 305-308 (1999). 28. Briant, D.J., Hankins, J.S., Cook, M.A. and Mackie, GA. The quaternary structure of RNase G from Escherichia coli. Mol. Microbiol. 50, 1381-1390 (2003). 29 Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structure of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105-124 (1993). 30 Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307-326 (1997). 31 Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772-1779 (2002). 32 Collaborative Computational Project No. 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760-763 (1994). 33 Cowtan, K. CCP4 ESF-EACBM Newslett. Protein Crystallogr. 31, 34-38 (1994). 34 Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022-1025 (1997). 35 Szöke, A., Szöke, H. & Somoza, J.R. Holographic Methods in X-ray Crystallography. CCP4 Daresbury Study Weekend Proceedings. 36 Emsley, P. & Cowtan, K. Coot: Model-Building tools for molecular graphics. Acta Crystallogr. D 60, 2126-2132 (2004). 37. Brunger, A.T. et al. Crystallography and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr. 54, 905-921 (1998). 38. Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D 53, 240-255 (1997). 8 39. Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Cryst. D 57, 122-133 (2000). 9