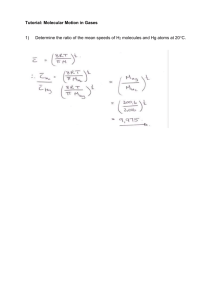
Here is where we stand:
advertisement

Here is where we stand: We have 2 different methods, (MSC and association). First to look at association: The probability of a domain pair interacting is equal to: While the probability of a protein pair interacting is equal to: The justification for these probabilities: Now to look at MSC: There are three different weight functions: 1- greedy algorithm using number of interactions 2- popularity using number of interactions 3- greedy algorithm using probabilities Now the probability here for a domain to interact is: While the probability of a protein pair interacting is equal to: Justification same as above? Now for the tables and metrics: For metrics: Specificity, sensitivity, number of matching p-p pairs, num p-p pairs predicted, number of p-p pairs tested (obtained experimentally), number of proteins pairs our training data set was composed of, total number of p-p pairs available in database, number of p-p pairs having a probability of interacting over some threshold. Problems: ChengBang and I disagreed on whether we should sum up the columns etc…. For example he has every percentage with is corresponding numbers, I suggest that it should be ranges, ex > 90%. I need to specifically state the scientific question within the context of the field of study, and its impact on computational biology. We also need to come up with a plan to test: We need composed data set and disjoint data set, (whether or not the training data set is included in testing data set). More detail is written in paper. We also need to decide whether we update the data structure or not once we choose max, for the three different weight functions we have. Work in Progress: As of now we are looking at other weight functions and finding different methods to compare them. One comparison to be made is to see which weight function needs the smallest number of domain pairs to explain the data. Another comparison can be made by seeing how many positive, negative, false positive, and false negative p-p interactions can be predicted in a set of data other than our training data and comparing these various numbers. Now this new set of data could be composed of our training data, disjoint, related to our training data, or not. -A composed data set would imply that learning data set is included among other observed protein interactions. -A disjoint data set implies that none of the protein interactions used in the training data are present in this new data set. -A related data set to the training data set could be p-p interactions observed in the same type of biological organism, same organ etc... - A unrelated data set could just be a different data set composed of p-p observations made from a different type of organism, different organ etc... - We also need to test on different sizes of training and observed data. If they are composed sets we just very training data size, otherwise we need to very both. We also plan on comparing our results with different results of existing programs by replicating their method for results using our algorithm. Some of the papers we plan on comparing with are: .Paper .Paper .Paper "Need to figure out which papers we are comparing with" . etc... .Paper The p-p database that we have been testing our different weight functions is composed of approximately 2300 p-p interactions. There exists a more up to date version of this data base which consists of approximately 12000 p-p interactions. We will be testing on this database pending an increase in the size of my AFS space and some optimization on the data structures in the algorithm, if the present algorithm is taking too much time to run. Some other weight functions that could be implemented: Weight function 5: As of now there are a lot of zeros present in our matrix. This is due to the fact that there are allot of different domains but most proteins only contain a few of these domains. I propose we take into account the absence of p-p interactions by introducing a negative weight. Yes this is similar to weight functions 3 and 4, but those weight functions calculate probabilities. With plain addition and subtraction I believe the algorithm will run much faster and the enormous number of zeroes in our matrix will come into play by becoming negative numbers. Besides the benefit of speed over the third and fourth weight function you would be able to give either the existence or the absence of a p-p edge more or less weight. Which might be desirable depending whether you believe you data set contains more false positives or is just simply incomplete. Plan of Attack (A program which does the calculations and prediction) First a list of what needs to be done: 1- A program which does the following: a. Reads in P-P interactions and stores them in a data structure (training data). An example input file: P1 P1 P1 P1 P2 P3 P4 P1 P2 P4 P5 P3 P5 P4 P4 P5 Now each protein will have a unique name and we can give each an ID number which is just some number being incremented every time a new protein is read in. So we would want a fast and efficient way to go from a proteins name no its ID. A protein should also know all the proteins it interacts with. And maybe store this number too. And it should know all the domains it contains which get read in from another file see c and maybe store this number too. The data structure will be a linked list of linked lists where the first one is all the proteins we re still looking at and the other is the proteins the protein interacts with. b. Reads in P-P interactions and stores them in a data structure (for testing purposes). Except we would probably need less info but for now leave it like a. All we really need to know here are all the proteins present in this testing case so we can predict the interactions and compare them to the experimental interactions. c. Reads in Protein structure and stores them in a data structure. An example of an input file is: >P1 D1 D3 // >P2 D1 D2 // >P3 D3 // >P4 D1 D4 // >P5 D5 // we also need to remove any p-p interactions which has a protein which we don’t know its structure and we should same it somewhere so we find out how many were removed d. Create a data structure containing the number/which proteins are contained in a domain. Kind of like a but now from domains, we want a domain id, proteins containing this domain, number of them, number of these proteins observed interacting, a list of which domain this domain could possibly be interacting with. e. Using the first 2 data structures created, create a third data structures containing information about possible D-D interactions. Already hinted at this in part d. We explain more about this data structure in a few: f. Create the different weight functions. (We have 4 of them now) g. These weight functions will be manipulating the data structure some slightly some will be changing entries to percentages etc…. and they will decide which is the best choice each time they are called h. Choose d-d pairs given weight function till all the p-p interactions are explained. Keep calling the specific weight function till there are no more p-p interactions in the testing data to be explained. i. Now time for predicting: We will take one domain pair at a time and see how many proteins pairs from the testing data contains this domain pair and we will predict that this pair of proteins interacts. Then we can check to see if they actually interact or not and look at j to see which numbers to increase etc… We keep doing this until we have went through all the domain pairs from our training data or all the proteins have already been explained. j. Use this set of d-d interactions to see how much of the p-p interactions from the testing data can be explained. i. How many could be explained, positives. ii. How many were predicted to interact but not observed to, false positives. iii. How many were not predicted to interact however they were observed as interacting (containing proteins with domains already studied), false negative. iv. How many were not predicted to interact however they were observed as interacting (containing proteins we don’t have any info about/ they have domains not observed in our training data). 2- 34- 5- v. How many were not predicted to interact and also not observed to interact. vi. Number of p-p pairs predicted which are over some threshold. vii. Calculate Specificity and Sensitivity. Test the program a. By creating small examples that I will follow through by hand to make sure it is doing what it is supposed. b. Testing on bigger data sets to get some results for comparison. Compare our results with what is already out there (publications related to this topic) Make final touches to paper, which I would have already been working on as I progress from one stage to the next. (Already about 4/5 pages including introduction and a brief explanation of the different weight functions.) Submit to a journal, conference or some publication.