MDEFT sequence for Siemens Trio
advertisement

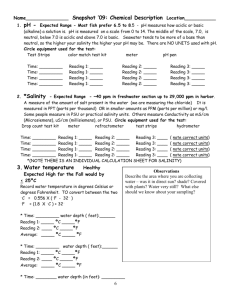
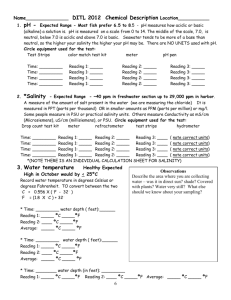
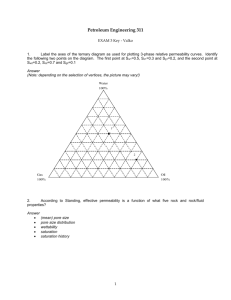
MDEFT sequence for Siemens Sonata Sequence Description The sequence ralf_mdeft_distrib_sonata is a 3D MDEFT sequence for T1 weighted structural brain imaging with whole brain coverage and an isotropic resolution of 1 mm. It should be used on Siemens 1.5T SONATA systems, only. The duration of the sequence is 12min. For a detailed description, please refer to: Deichmann et al., NeuroImage 21, 757-767 (2004) Implementation Please compile the source code and put the binary files into the appropriate directory on the scanner's host computer. When you create the protocol for the sequence: Most of the parameters will already have the correct setting. Basically, only the following changes should be necessary in POET: 1. In ROUTINE: Change the slab orientation to sagittal (Leave phase encoding on A>>P) 2. Make sure the correct coil is selected 3. There is a 5cm transverse saturation bar. This is used for the saturation of inflowing blood (to avoid flow artefacts) and should be located directly below the cerebellum without saturating parts of the brain. My recommendation is to change in GEOMETRY/SATURATION the position of the saturation slab to 100 mm in feet direction (i.e. the position is L0.0 P0.0 F100.0) Please note: on Sonata and Trio systems, usually a body coil is used for excitation. In this case, saturation of inflowing blood should not be necessary and the saturation bar can be switched off. Sequence parameters In detail, the parameters in the different task cards are: ROUTINE: sagittal, phase enc. in A>>P, 176 slices per slab, FoV=256mm, FoV=87.5% in phase, TR/TE=12.24/3.56 CONTRAST: TI=530, flip angle=23deg RESOLUTION: slice thickness=1mm, base resolution=256, phase res. =100%, no filters selected GEOMETRY/SATURATION: transverse saturation, 50mm, position L0.0 P0.0 F100.0 SEQUENCE/PART1: BW=106 HZ/Pixel SEQUENCE/PART2: RF spoiling ON SEQUENCE/SPECIAL: saturation angle:110deg (for blood suppression), relaxation delay=0, first part of TI: 42%, FATSAT-angle=160deg Use of sequence When planning the imaging slab on the scout scan: use parallel shifts only, please avoid rotations. For subjects with big heads, the nose might cause aliasing artefacts. In his case, you have to increase the FOV in phase encoding direction. Please do so by changing in the Resolution task card the entry FoVPhase from 87.5% to 93.8% or even 100%. However, this will increase the duration to 12min 51sec or 13min 43sec. Remarks and Warnings 1. Please test the sequence on a phantom first. Please check the SAR values during the phantom experiment. 2. Please make sure you stick to the parameters listed above. Even if some parameters can be changed on the interface, it is likely that this reduces the image quality. As an example, the change of the number of slices per slab and/or of the flip angle may result in blurring. Safety relevant figures SAR Typical SAR values are: 0.19 W/kg (head), 0.12 W/kg (exposed body), 0.06 W/kg (whole body). Values were obtained for female subject with body weight of 75 kg dB/dt The slew rate of the read gradient for the parameters given above is 91.7 mT/m/ms This means that at a position inside the gradient coil which has a distance of 10 cm from the read axis (head/foot axis), dB/dt is 9.17 T/s. Test report The sequence code was tested by creating a protocol ralf_mdeft_distrib_sonata.pro using the sequence parameters given above, and running the Siemens unit test: ut -bp ralf_mdeft_distrib_sonata.pro Please refer to the attached file unit_test_sonata.doc for test results. Due to the use of special excitation pulses that compensate the effects of B1 inhomogeneities and require gradient pulses in different directions during RF transmission, the following unit test components were disabled: - Incorrect slice-select gradient amplitude (RFAmplValErr) - Slice-select gradient (RTEIDENT_PulseSS) is not in the slice-select channel (GrNotInRoRFErr) - Slice thickness defined in RF pulse structure not the same as protocol value (WrongSliceThickErr) The sequence passed the unit test. Two features of the test resulted in "Not OK": - Image orientation - Slice select gradient amplitude This is due to disabling the test features listed above. To check for correct image orientation, the sequence was run on a healthy volunteer and the resulting data set was compared to images acquired with a Siemens release sequence. Result: image orientations were identical, left and right were not swapped. Disclaimer Although we have done our best to assess safety issues, we cannot exclude the risk of sequence malfunctions or the occurrence of safety-related problems. It is mutually agreed that the sequence is handed over on the basis of a research cooperation and that the respective institution accepts the following disclaimer: ######################################################################## The MR sequence was designed for research purposes only. The developer(s) of the sequence cannot assume any responsibility for problems arising from its use. This applies specifically to the cases of: - Damage to subjects - Damage to hardware - False diagnosis and wrong decisions based on data acquired with the sequence It is mutually agreed that the sequence code is to be considered a template only, and that it is the responsibility of the MR physicist who modifies and compiles the code and installs the binaries on site to run the system-specific tests and assess security and safety issues. It is mutually agreed that the sequence was designed for research purposes only and is handed over on a non-profit basis. ########################################################################