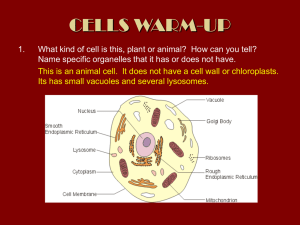
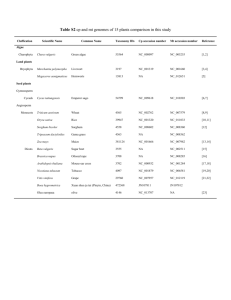
In silico identification of conserved proteins in sequenced
advertisement

In silico identification of conserved proteins in sequenced chloroplast genomes of C3 plants Dhwani Raghav, Asheesh Shanker and Vinay Sharma Department of Bioscience and Biotechnology Banasthali University, Rajasthan-304022 Corresponding author’s Email: sasheesh@bv.banasthali.ac.in Abstract Chloroplasts are the organelles that capture light energy to conserve it in the form of ATP and reduce NADP to NADPH through a complex set of processes called photosynthesis. The chloroplast genome was the first plant genome characterized. A large number of databases (NCBI, EMBL, DDBJ) were created to store sequences of various genomes.These databases also contains tools to analyze these sequences. Using BASIC LOCAL ALIGNMENT SEARCH TOOL (BLAST), conserved and unique proteins between different species of C3 plants, namely Atropa belladonna, Cucumis sativus, Glycine max, Gossypium hirsutum and Lotus japonicus were identified. Conserved proteins play an important role in phylogenetic analysis, while unique proteins provide a new shed of light in the field of functional and comparative genomics. 1. INTRODUCTION The chloroplast is the hallmark organelle of plants (Gr. Chloros means green and plastos means formed). These specialized organelles are one of the many unique cells in the body and are generally considered to be originated endosymbiotically [12]. Chloroplasts contain the chlorophyll, hence provide the green color and used for critical functions such as photosynthesis, starch synthesis, nitrogen metabolism, sulfate reduction, fatty acid synthesis, DNA and RNA synthesis [17], and are also involved in the energetic processes in plants [9]. Chloroplasts have their own DNA which resembles large bacterial plasmids or small chromosomes and it inherited cytoplasmically, independent of nuclear genes [16]. Its small size ranges from about 120-220 kb with 120-150 genes [14] and limited number of repeated elements made it a prime candidate for characterization. The gene content and arrangement of chloroplast DNAs (cpDNAs) in higher plants are relatively uniform from species to species. Its abundance in foliar tissue made it easy to isolate. The inverted repeat is an interesting feature of the chloroplast genome. Completely sequenced chloroplast genomes provide a rich source of data that can be used to find phylogenetic relationship [4]. C3 plants are those in which C3 carbon fixation takes place, which is a pathway for carbon fixation in photosynthesis. In this process carbon dioxide and ribulose bisphosphate (RuBP, a 6carbon sugar) is converted into phosphoglycerate, which is a 3- carbon compound. Melvin Calvin and his coworkers at the University of California worked out the mechanism of the reduction of CO2 and suggested cyclic pathways where the first stable compound produced during carbon reduction is a 3-C compound, i.e., 3-phosphoglyceric acid. In C3 plants, carbon fixation and photosynthesis happens in mesophyll cells just on the surface of the leaf. Using bioinformatic tools to compare important species is now feasible and structural genomics continue in importance and establishment of structure-function relationships become a common way of comparative analysis. Inference of relationships from proteins of known function to proteins of unknown function that are structurally similar can be established through comparative analysis [6]. The smallness in size of chloroplast genomes facilitates for the discovery of disease resistance genes, introgression of important traits in transgenic plants quantitative trait analysis [11], and phylogenetic studies [4] etc. The Chloroplast Genome Database (CGD; ChloroplastDB) [3] is an interactive, web-based database for fully sequenced plastid genomes, containing genomic, protein, DNA and RNA