SafePGR
advertisement

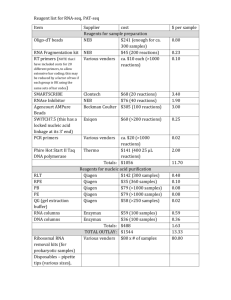
SafePGR Videonference report 26/09/2012 L. Bonheur, T. Candresse, M. Carvalho, D. Filloux, M. Grisoni, C. Julian, D. Mendonça, C. Pavis, T. Santos, E. Silva, P.-Y. Teycheney Workpackage 1, Viral diversity Follow-up of plant samplings, TNA extractions and PCR is summarized in an Excel file to be made available to participants through Dropbox . o La Réunion has started analyses on garlic; 40 samples analysed; viruses identified, sequences generated for genetic diversity study. Further phylogenetic analyses may be performed for a publication. Difficulties for extracting TNA from sweet potato and yam were encountered when using the simplified protocol provided by INRA Bordeaux: coloured extracts, due to oxidation or other, no amplification following PCR with housekeeping gene primers. Tried CTAB, Trizol and Qiagen on fresh or lyopphylized leaves. Best results with Qiagen Plant DNeasy minikit when using twice less plant material than what supplier recommends. o Montpelier recommends to use Qiagen Plant DNEasy minikit for extracting total DNA from yam, and double the number of rinses prior to elution step. o Guadeloupe has hired Lydiane Bonheur in September. Sampling of banana and sugarcane has started and is complete for yam. No sweet potato samples yet. Waiting for missing reagents to start TNA extractions. o Azores : tried TNA extraction from sweet potato using the simplified protocol provided by INRA Bordeaux ; very average quality when checked by Nanodrop and electrophoresis o Madeira : didn’t start extracting TNAs yet because of delays in getting reagents. Will start next week with Armelle. Primer design for new species identified from EST analyses: see below (WP3) Guadeloupe and Azores to test DNA extraction protocol on sweet potato and yam, then report to other participants A shared Excel file will be put by Claudie in a Dropbox folder, to follow-up task 1 of WP1. Workpackage 2, Metagenomics Charlotte started to extract small RNAs on 6 species and sent them for sequencing. Waiting for sequencing results. Finished the semi purification of viral particles from 6 species (16 samples / species). Started the double strand RNA extraction, which proves difficult on yam , with very low yields and poor quality. Similar problems were encountered by Lydiane in Guadeloupe. Guadeloupe sent infected leaf material (potexvirus, sadwavirus) to Bordeaux, who will extract dsDNA and send them back to Guadeloupe for PCR analyses. Workpackage 3, Bioinformatics Data mining is now complete, with results almost completely described in the Thierry’s 6 month report. This report will be completed when results will be available for all primers. o Yam closterovirus (1 EST): Thierry will design a primer pair o Banana tobamovirus (1 EST): Thierry will design a primer pair o Sugarcane betaflexivirus (1 EST) : Philippe & Denis to decide whether it is worth designer primers o Sweet potato varicosavirus / cytorhabdovirus : Denis to design primers; wil be tested once TNA extraction is OK o Mikania micrantha mosaic virus (Fabavirus) in the sweet potato illumina contigs (5 contigs in total): not worth loking, if widespread in sweet potato, then it will come out from the deep sequencing data o Garlic foveavirus: several ESTs of a new foveavirus for which primers were designed and used on TNAs; analyses show that the virus is present and widespread in Réunion garlic samples, with some diversity among strains; most of the genome is covered by ESTs. o Yam macluravirus : 5 primers have been designed by Denis and tested on D. alata positive controls provided by S. Winter, with good results. Denis awaiting additional samples originating from China, to be provided by a Japanese team, for confirming results, then he will send protocol to partners. o Sugarcane mastrevirus: primers have been designed by Denis Workpackage Management The 6-month report has been sent on time to the NetBiome JCS and to the funders D3.1 & D3.2 are achieved. Will have to be completed/consolidated once primers have been designed and sent to Guadeloupe Other deliverables have to be formalized when ready (screening of crops): ESTs for viral sequences, sequence information on novel viral agents to WP1 (1/10/2012). Next videoconference : first week of December Next project meeting in Montpellier around 12-20 September Claudie prepares Doodle surveys for both events.