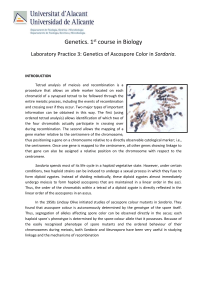
Analyzing the progeny of an outcrossed cleistothecium
advertisement

Ex4 Genetic analysis in Aspergillus nidulans. page 1 of 2 We have been studying fungal growth and response to changing environments, and mold isolation and identification. With few exceptions, none of the organisms in these lab exercises were characterized beyond genus. In contrast, when fungi are used as experimental tools they are generally very precisely identified and characterized. The matings for this lab were done between strains with different spore colours and nutritional auxotrophies (see table below). In order to test whether individual cleistothecia resulted from an outcross, that is, between parental strains, a small volume of ascospore suspension is spread on rich medium and incubated at 28°C for two to three days. The ascospore suspension is stored at 4°C, where the spores will remain viable for months to years. Today you’ll be cleaning Aspergillus cleistothecia, suspending the ascospores in sterile distilled water, and plating a sample of spores to test whether they came from a cross between the two parents. Aspergillus can mate with itself, so this is not a trivial test. However, we will pick cleistothecia that are likely to be outcrossed by choosing large ones formed close to conidia that have both parental spore colours. The conidia will be plated on medium that will support most of the auxotrophies, enough that we will be able to tell outcrosses for next time. Analyzing the progeny of an outcrossed cleistothecium Just learning that you have an outcrossed cleistothecium doesn’t get you very far. Generally you are trying to do two things – learn about a genetic interaction which sometimes requires isolating strains that have particular combinations of mutations, and determining the nutritional requirements of those strains. To accomplish these aims, you need single-spore progeny. The original spread of your newly isolated ascospores was done at high spore density. All you needed then was to distinguish between outcrossed and self-crossed cleistothecia. The next objective requires precision: you will be spreading a diluted spore sample on more CM+ plates, and next time you will be picking those progeny for analysis. Preparation: make a glass spreader. Get a 5 1/4 inch Pasteur pipette. Hold the narrow end in a Bunsen burner flame until it seals and droops slightly. Pull the pipette out of the flame, and rotate it about 90° until the droopy tip is pointing towards you. Then, flame again so that the pipette bends about an inch from the tip. Remove and place in an Eppendorf rack to cool. Ex4 Genetic analysis in Aspergillus nidulans. page 2 of 2 Examine the Eppendorf tubes that have your ascospore suspension: you should be able to see a dark spot at the bottom, which is the mass of ascospores originally in the cleistothecium. Obviously, these spores are heavier than water. To spread plates for isolated colonies (likely to have been produced from a single spore), thoroughly mix your original ascospore suspension and make a 1:100 dilution in sterile distilled water. Use 2µl of ascospores in 200 µl of water. Mix again, and them pipette 30µl, 60µl and 90µl of this suspension onto three labeled plates of CM+ agar. Spread the suspension evenly over the surface of the plate using a sterile spreader. Dip your spreader in alcohol, flame briefly, dip again in alcohol, flame to dry and let cool. Test the coolness by touching the agar. Leave the plates right side up for 30-60 min to allow the water in the suspension to absorb into the agar. Tape shut and invert. Store at room temperature for two to three days. A. nidulans ascospores take longer to germinate than do conidia. We will be analyzing individual ascospore progeny by random ascospore analysis. Colonies from single ascospores are sampled by probing gently with a sterile toothpick that was first by touching to a clean part of the agar, and then point inoculated by touching to a master plate that has a fixed position with respect to the locator grid. Each toothpick is used for only one colony, but several replicate plates can be inolculated from the same toothpick. This whole process must be done with minimal air disturbance. The plates are opened once only for the entire process, you should avoid waving your hands over the source or recipient plates, and you should not breathe over the plates. Done properly, you will have 50 progeny plus 2 parental colonies growing well separated on a single plate. Otherwise, you will have many small colonies as well as the ones you inoculated intentionally. By inoculating replicate series of colonies onto media of different compositions, or grown under different conditions, you will be able to deduce the genotype of each progeny strain. It is important to inoculate the two parental strains onto each plate for cross reference purposes. The parental strains have been well characterized, but some of the phenotypes of the alleles can be difficult to score. Generally the strains mated for this cross have easy to score phenotypes. Scoring phenotypes used sheets designed for convenient tallying of characteristics. An interesting analysis is to compare inheritance amongst pairs of genes. You might find that if the parental cross is AB :: ab, that these combinations occur far more frequently than then recombinant Ab and aB. This can be tested statistically to determine if the genes are linked, that is, whether they are physically close on the same chromosome. Regardless of linkage (which is only relevant when genes are compared pairwise), a/A and b/B should be inherited by 50% of the progeny. Your tasks for this exercise will be to identify and isolate strains that have interesting and useful genetic marker comparisons for future research to study gene linkage to study gene epistasis, using colour markers