Thesis - Université Paris
advertisement

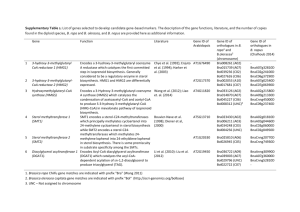
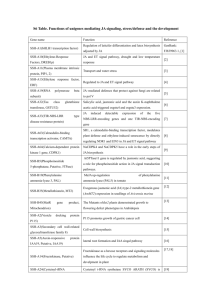
PhD Thesis proposal form Biology Doctoral School 426 : Gènes, Génomes, cellules Thesis subject title: New pyrrolamide antibiotics by combinatorial biosynthesis and mutasynthesis Laboratory name and web site: Molecular Microbiology of Actinomycetes, http://www.igmors.u-psud.fr/spip.php?rubrique195&lang=en PhD supervisor (contact person): Name: Dr Jean-Luc Pernodet Position: Team leader email: jean-luc.pernodet@igmors.u-psud.fr Phone number:+33 1 69 15 46 41 Thesis proposal (max 1500 words): Context of the project Natural products have long constituted the sole source of human remedies and medicines and nowadays, they still occupy a preponderant place in our pharmacopoeia, notably as antimicrobial and anticancer drugs. Indeed, using newly developed methods (genome mining) and techniques (automated dereplication of natural products and high throughput screening), an active search is conducted worldwide to uncover new microbial natural products with pharmaceutical potential. Parallel to these efforts, new approaches have recently emerged to create new “unnatural” natural products. Two main strategies, which are not exclusive of each other, are being explored: combinatorial biosynthesis and mutasynthesis. Combinatorial biosynthesis uses original combinations of biosynthetic genes from various natural product clusters to generate natural product analogues. Mutasynthesis is based on the feeding of microbial strains, blocked in the biosynthesis of an intermediate, with chemically synthesized intermediate analogues. The resulting products are hybrids between the artificial building block and the original natural compounds. These approaches have already been successful, yielding, for example, aminocoumarin derivatives with higher antibacterial activities than the clinically used novobiocin. This project aims at developing a combinatorial biosynthesis- and mutasynthesis-based approach for the development of new bioactive molecules. It is centered on the pyrrolamide antibiotic family, well studied in the host laboratory. Pyrrolamides (e.g. congocidine, distamycin, kikumycins, pyrronamycins, noformycin), constitute a family of natural products produced by Streptomyces or related actinobacteria. They exhibit a variety of biological activities, such as antiviral, antibacterial, antitumor and anthelmintic activities. Figure 1: Structures of A) congocidine Isolated about sixty years ago, the two best characterised and B) distamycin. Doted lines are used members of the pyrrolamide family, congocidine (also called netropsin) and distamycin (Figure 1), have been to separate putative precursors. extensively studied due to their ability to bind to the minor groove of the DNA double helix in a sequence-specific manner (Neidle, 2001; Dyatkina et al, 2002; Khalaf et al., 2004). They serve as models for the study of DNA minor-groove binders and it has been shown that they bind DNA reversibly at sequences of four or more consecutive A-T pairs and strongly discriminate against G-C pairs (Neidle, 2001). These molecules are still the focus of substantial attention and many analogues and hybrids of congocidine and distamycin have been synthesised chemically in attempts to modify or increase the sequence specificity of the DNA binding (Neidle, 2001), or to create new nontoxic antimicrobial molecules (Dyatkina et al, 2002, Khalaf et al., 2004, Anthony et al. 2007). Pyrrolamide gene clusters studied in the host laboratory In the laboratory, we characterized the first gene cluster directing the biosynthesis of a pyrrolamide, congocidine, in Streptomyces ambofaciens (Juguet et al., 2009)(Figure 2). We showed that congocidine is assembled by a nonribosomal peptide synthetase (NRPS), constituted of Cgc2, Cgc16, Cgc18 and Cgc19, that displays some unusual features. Its single adenylation domain acts iteratively and one of its condensation domain uses CoA- rather than PCP-Ppant-activated guanidinoacetate as a substrate. Moreover, it is constituted of a module and domains that are freestanding. This organisation and the model we propose for the assembly and tailoring of the congocidine backbone seems particularly suited for the assembly of poly-pyrrole-2-carboxamides such as congocidine (two pyrrole-2-carboxamides) or distamycin (three pyrrole-2-carboxamides) and makes of pyrrolamides appealing candidates for the development of a combinatorial biosynthesis approach. Figure 2: Organisation of the gene cluster directing the biosynthesis of congocidine in Streptomyces ambofaciens. In addition, we recently established the nature of the three precursors assembled by the congocidine NRPS. We showed that Cgc7, an enzyme that bears no resemblance with proteins in databases is involved in the guanidinoacetate precursor biosynthesis (Lautru et al. (a), in preparation). We established the biosynthetic pathway to 3-amidinopropionamidine from cytidine monophosphate through a combination of functional analyses of cgc genes and chemical complementations and showed that it involves the three enzymes Cgc4, Cgc5 and Cgc6 (Lautru et al. (a), in preparation). Finally, we identified 4-acetamidopyrrole-2-carboxylate as the key congocidine pyrrole precursor and proposed a biosynthetic pathway to it from N-acetylglucosamine (Lautru et al. (b), submitted to Angewandte Chemie). We showed that five enzymes encoded within the congocidine biosynthetic gene cluster with sequence similarity to known carbohydrate-processing enzymes participate in this pathway, which bears no resemblance to other pyrrole biosynthetic pathways. To develop a combinatorial biosynthesis approach for the development of new biologically active pyrrolamide derivatives, it is necessary to have at hand several characterised biosynthetic gene clusters directing the biosynthesis of compounds of the same family. We have therefore identified and isolated gene clusters directing the biosynthesis of two other pyrrolamides, distamycin and pyrronamycins. Distamycin is struturally closely related to congocidine but possesses three pyrrole rings. The differences in the structure of pyrronamycins and congocidine/distamycin suggest that the characterization of its biosynthesis are is likely to bring new tools (genes/enzymes) and bricks (precursors) necessary to a succesful combinatorial biosynthesis approach. PhD work program 1. Functional analysis of the distamycin and pyrronamycin gene clusters The first part of the project will consist in establishing the biosynthetic steps specific to distamycin or to pyrronamycins syntheses. For this purpose, gene deletions and LC-MS analyses of culture supernatant of the resulting mutant strains will be carried out. Potential intermediates accumulating in the culture medium will be purified and characterized in collaboration with chemists. If possible, chemical complementation with synthesized intermediates or precursors will be attempted. This analysis will result in a fine understanding of the mechanisms underlying distamycin and pyrronamycins biosyntheses, necessary to rationally design combinatorial biosynthesis and mutasynthesis experiments for the synthesis of new pyrrolamides derivatives. 2. Synthesis of new pyrrolamide derivatives by combinatorial biosynthesis One of the first objectives of the combinatorial biosynthesis experiments will be to vary the number of pyrrole precursors incorporated in the final molecule. This will be achieved by exchanging genes encoding the nonribosomal peptide synthetase domains or modules between the different pyrrolamide gene clusters. The next step will consist in varying the nature of the precursors other than the pyrrole groups to be incorporated in the final molecule. Indeed, apart from the pyrrole groups common to all pyrrolamides, five distinct groups can be identified in the structures of congocidine, distamycin and pyrronamycins: the guanidinoacetyl group (congocidine and pyrronamycins), the 3aminopropionamidyl group (congocidine and distamycin), the formyl group (distamycin) and the 3amino-3-cyano propionic acid and 2,6-diamino-4-hexenal in pyrronamycins. Precursor biosynthesis gene cassettes will be carefully designed to be used for the creation of custom-made pyrrolamide derivatives. Finally, to add further chemical complexity to the new pyrrolamide derivatives obtained in the first two steps, genes involved in the tailoring of the pyrrolamide molecules (for e.g., methyltransferase or guanidinoacetate hydroxylase genes) will be added to the newly constructed gene clusters. Pyrrolamide derivatives obtained by combinatorial biosynthesis will be tested for their biological activities, thus providing information on structure-activity relationships. 3. Synthesis of new pyrrolamide derivatives by mutasynthesis This part of the project will consist in feeding to various mutant strains blocked in the biosynthesis of one or several pyrrolamide precursors a number of chemically synthesized unnatural precursors to determine whether these precursors can be incorporated by pyrrolamide biosynthetic systems. This will be carried out in collaboration with the group of Dr Laurent Micouin (University Paris Descartes) who will provide the unnatural precursors to be fed to the various mutant strains. Pyrrolamide derivatives obtained by mutasynthesis will be tested for their biological activities. This PhD project is part of a broader multidisciplinary research project for which a grant proposal (involving the host laboratory and the laboratory of Dr Laurent Micouin, University Paris Descartes) has been deposited for funding by the French ANR. The PhD student involved in this project will therefore be part of a team working on combinatorial biosynthesis and mutasynthesis. He will have the opportunity to work in a multidisciplinary environment and to acquire skills/expertise in microbiology, molecular biology and analytical chemistry and to contribute to the development of new tools for combinatorial biosynthesis and synthetic biology in the field of natural products. References Anthony N.G., Breen D. et al. (2007) J. Med. Chem. 50: 6116-6125 Dyatkina N.B., Roberts C.D. et al. (2002) J. Med. Chem. 45: 805-817. Juguet, M., Lautru, S. et al (2009) Chemistry & Biology 16: 421-431. Khalaf A.I., Ebrahimabadi A.H. et al. (2004). Org. Biomol. Chem. 2: 3119-3127 Lautru S., Malet N., et al. (a) In preparation. Lautru S., Song L. et al (b) A sweet origin for the key congocidine precursor 4acetamidopyrrole-2-carboxylate. Submitted to Angewandte Chemie International Edition. Neidle S. (2001). Nat. Prod. Rep., 18: 291-309. Publications of the laboratory in the field (max 5): 1. Lautru S. Song L., Demange L., Lombès, T., Galons, H, Challis G.L. and Pernodet J-L. A sweet origin for the key congocidine precursor 4-acetamidopyrrole-2-carboxylate. Submitted to Angewandte Chemie. 2. Seguin J, Moutiez M, Li Y, Belin P, Lecoq A, Fonvielle M, Charbonnier JB, Pernodet JL, Gondry M (2011), Nonribosomal peptide synthesis in animals : the cyclodipeptide synthase of Nematostella, Chemistry and Biology, 18(11):1362-1368 3. Nguyen HC, Karray F, Lautru S, Gagnat J, Lebrihi A, Huynh TD and Pernodet JL, (2010), Glycosylation steps during spiramycin biosynthesis in Streptomyces ambofaciens : involvement of three glycosyltransferases and their interplay with two auxiliary proteins, Antimicrob Agents Chemother, 54(7):2830-9 4. Juguet M, Lautru S, Francou F-X, Nezbedová S, Leblond P, Gondry M and Pernodet J-L. (2009) An iterative nonribosomal peptide synthetase assembles the pyrrole-amide antibiotic congocidine (netropsin) in Streptomyces ambofaciens. Chemistry and Biology,16 : 421-431 5. Gondry M., Sauguet L., Belin P., Thai R., Amourouc R., Tellier C, Tuphile K, Jacquet M, Braud Scourçon M, Masson C., Dubois S, Lautru S, Lecoq A., Hashimoto SI, Genet R and Pernodet J.-L. (2009) Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nature Chemical Biology, 5 : 414-420. Specific requirements to apply, if any: The candidate should have a background in microbiology and molecular biology. Basic knowledge in organic and analytical chemistry will be an advantage.