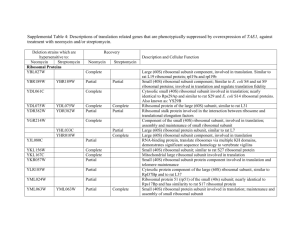
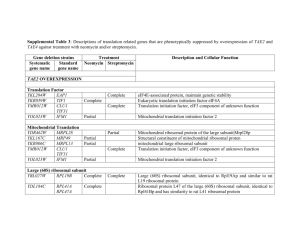
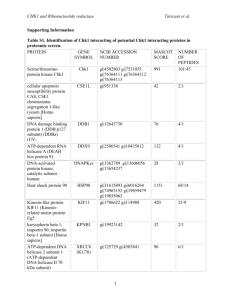
Proteomic screening identifies protein aggregation as the
advertisement

Anti-amyloid compounds protect from silica nanoparticle-induced neurotoxicity in the nematode C. elegans Andrea Scharf, Karl-Heinz Gührs, Anna von Mikecz Supplemental Material Figure S1. Representative fluorescent micrographs of 2-day-old (young adult) C. elegans stably expressing (A) a homopolymeric glutamine repeat fused to yellow fluorescent protein (Q24::YFP), (B) Q35::YFP, and (C) Q40::YFP under the control of the unc-54 promoter in the body wall muscle cells. Worms were mock (H2O)- or silica NP-treated in liquid culture for 24 hours. Bar, 100 µm. Q24, Q35 or Q40, homopolymeric repeats of 24, 35 or 40 glutamines, respectively; YFP, yellow fluorescent protein. Figure S2. Gene expression of proteasomes in silica NP-exposed worms. (A) 20S proteasome expression levels of total protein extracts of 2-day-old, adult wild-type C. elegans that were mock- (H2O) or silica NP-treated for 24 hours. Worms were lysed in SDS-lysis buffer (Scharf et al., 2013), proteins were separated via SDS-polyacrylamide-gelelectrophoresis, and analyzed by immunoblotting with antibodies against the alpha-subunits of the 20S proteasome. Separated proteins were stained with Coomassie Brilliant Blue as loading control. (B) Respective densitometric quantification of the immunoblot in (A). Values represent means +/- SD. A, anti; kDA, kilodalton; MW,molecular weight. Figure S3. Intact morphology of body wall muscles after exposure of C. elegans with silica NPs. Fluorescence microscopy of a representative 3-day-old (young adult) worm stably expressing green fluorescent protein (GFP) under the control of the myosin-3 (myo-3) promoter that was treated with rhodamine-labelled silica NPs for 48 hours. Arrowheads indicate intact sarcomere structure, e.g. sarcomere banding, in body wall muscles. Bar, 10 µm. Table S1. Control C. elegans aggregome as identified by mass spectrometry analysis. entry name protein names translation, rRNA metabolic process RS14_CAEEL 40S ribosomal protein S14 R23A2_CAEEL 60S ribosomal protein L23a 2 GBLP_CAEEL Guanine nucleotide-binding protein subunit beta-2-like 1 metabolic processes PYC1_CAEEL Pyruvate carboxylase 1 (Pyruvic carboxylase 1) (PCB 1) ENO_CAEEL Enolase (2-phospho-D-glycerate hydro-lyase) (2-phosphoglycerate dehydratase) ion transport AT1B1_CAEEL Sodium/potassium-transporting ATPase subunit beta-1 VDAC_CAEEL Probable voltage-dependent anion-selective channel transport FOLT2_CAEEL Folate-like transporter 2 others OST3_CAEEL Probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3 Table S1 lists all filter-trapped SDS-insoluble proteins specifically detected in H2O-treated 2day-old, adult wild-type C. elegans. Candidates were characterized by mass spectrometry and the measured spectra were searched against the SwissProt database by Mascot. Entry names are according to SwissProt and UniProt databases. Candidates were detected in at least two from four independent experiments. Table S2. The intersection–aggregome of H2O- and silica NP-treated C. elegans as identified by mass spectrometry analysis. entry name protein names translation, rRNA metabolic process CGH1_CAEEL ATP-dependent RNA helicase cgh-1 (EC 3.6.4.13) (Conserved germline helicase 1) RL10_CAEEL 60S ribosomal protein L10 (QM protein homolog) RL10A_CAEEL 60S ribosomal protein L10a RL12_CAEEL 60S ribosomal protein L12 RL13A_CAEEL 60S ribosomal protein L13a RL19_CAEEL 60S ribosomal protein L19 RL23_CAEEL 60S ribosomal protein L23 RL3_CAEEL 60S ribosomal protein L3 RL35_CAEEL 60S ribosomal protein L35 RL4_CAEEL 60S ribosomal protein L4 RL5_CAEEL 60S ribosomal protein L5 RL6_CAEEL 60S ribosomal protein L6 RL7A_CAEEL 60S ribosomal protein L7a RS15_CAEEL 40S ribosomal protein S15 RS2_CAEEL 40S ribosomal protein S2 RS21_CAEEL 40S ribosomal protein S21 RS26_CAEEL 40S ribosomal protein S26 RS3_CAEEL 40S ribosomal protein S3 RS3A_CAEEL 40S ribosomal protein S3a RS8_CAEEL 40S ribosomal protein S8 RSSA_CAEEL 40S ribosomal protein SA SYV_CAEEL Valine--tRNA ligase (EC 6.1.1.9) (Valyl-tRNA synthetase) (ValRS) protein folding, proteolysis, stress response HSP7A_CAEEL Heat shock 70 kDa protein A HSP90_CAEEL Heat shock protein 90 (Abnormal dauer formation protein 21) TCPA_CAEEL T-complex protein 1 subunit alpha (TCP-1-alpha) (CCT-alpha) ERP1A_CAEEL Putative endoplasmic reticulum metallopeptidase 1-A (EC 3.4.-.-) (FXNA-like protease) muscle contraction MYO1_CAEEL Myosin-1 (Lethal protein 75) (Myosin heavy chain D) (MHC D) MYO2_CAEEL Myosin-2 (Myosin heavy chain C) (MHC C) MYO3_CAEEL Myosin-3 (Myosin heavy chain A) (MHC A) MYO4_CAEEL Myosin-4 (Myosin heavy chain B) (MHC B) (Uncoordinated protein 54) TBA2_CAEEL Tubulin alpha-2 chain TBB2_CAEEL Tubulin beta-2 chain (Beta-2-tubulin) UNC22_CAEEL Twitchin (EC 2.7.11.1) (Uncoordinated protein 22) UNC52_CAEEL Basement membrane proteoglycan (Perlecan homolog) (Uncoordinated protein 52) (Protein unc-52) UNC89_CAEEL Muscle M-line assembly protein unc-89 (Uncoordinated protein 89) metabolic processes 3HIDH_CAEEL Probable 3-hydroxyisobutyrate dehydrogenase, mitochondrial (HIBADH) (EC 1.1.1.31) ACOC_CAEEL ALF1_CAEEL Probable cytoplasmic aconitate hydratase (Aconitase) (EC 4.2.1.3) (Citrate hydro-lyase) (Gex-3-interacting protein 22) Fructose-bisphosphate aldolase 1 (EC 4.1.2.13) (Aldolase CE-1) (CE1) ECHM_CAEEL Probable enoyl-CoA hydratase, mitochondrial (EC 4.2.1.17) GCP_CAEEL Bifunctional glyoxylate cycle protein (Gex-3-interacting protein 7) [Includes: Isocitrate lyase (ICL) (Isocitrase) (Isocitratase) (EC 4.1.3.1); Malate synthase (EC 2.3.3.9)] Probable arginine kinase F46H5.3 (AK) (EC 2.7.3.3) KARG1_CAEE L LE767_CAEEL MPCP_CAEEL Very-long-chain 3-oxooacyl-coA reductase let-767 (EC 1.1.1.330) (Lethal protein 767) (Putative steroid dehydrogenase let-767) (Short-chain dehydrogenase 10) Probable S-adenosylmethionine synthase 1 (AdoMet synthase 1) (EC 2.5.1.6) (Methionine adenosyltransferase 1) (MAT 1) Probable methylmalonate-semialdehyde dehydrogenase [acylating], mitochondrial (MMSDH) (Malonatesemialdehyde dehydrogenase [acylating]) (EC 1.2.1.18) (EC 1.2.1.27) Phosphate carrier protein, mitochondrial (PTP) OAT_CAEEL Probable ornithine aminotransferase, mitochondrial (EC 2.6.1.13) (Ornithine--oxo-acid aminotransferase) ODO1_CAEEL 2-oxoglutarate dehydrogenase, mitochondrial (EC 1.2.4.2) (2-oxoglutarate dehydrogenase complex component E1) (OGDC-E1) (Alpha-ketoglutarate dehydrogenase) METK1_CAEE L MMSA_CAEEL ion transport ATPA_CAEEL ATP synthase subunit alpha, mitochondrial ATPB_CAEEL ATP synthase subunit beta, mitochondrial (EC 3.6.3.14) ATPL2_CAEEL Probable ATP synthase subunit g 2, mitochondrial (ATPase subunit g 2) P90735_CAEEL Protein EAT-6 G5EC39_CAEEL Calcium-transporting ATPase (EC 3.6.3.8) Q9TYP9_CAEEL Calcium-transporting ATPase (EC 3.6.3.8) Q95XP5_CAEEL Calcium-transporting ATPase (EC 3.6.3.8) G5EEK8_CAEEL Calcium-transporting ATPase (EC 3.6.3.8) VATA_CAEEL V-type proton ATPase catalytic subunit A (V-ATPase subunit A) (EC 3.6.3.14) (V-ATPase 69 kDa subunit) (Vacuolar H ATPase protein 13) (Vacuolar proton pump subunit alpha) Probable V-type proton ATPase subunit B (V-ATPase subunit B) (Vacuolar proton pump subunit B) VATB_CAEEL VATL2_CAEEL V-type proton ATPase 16 kDa proteolipid subunit 2/3 (V-ATPase 16 kDa proteolipid subunit 2/3) (Vacuolar proton pump 16 kDa proteolipid subunit 2/3) chromatin, structure H4_CAEEL Histone H4 storage VIT1_CAEEL Vitellogenin-1 VIT2_CAEEL Vitellogenin-2 VIT5_CAEEL Vitellogenin-5 VIT6_CAEEL Vitellogenin-6 transport CLH_CAEEL Probable clathrin heavy chain 1 Table S2 lists all filter-trapped SDS-insoluble proteins detected in both H2O- or silica NPtreated 2-day-old, adult wild-type C. elegans (intersection). Candidates were characterized by mass spectrometry and measured spectra were searched against the SwissProt database by Mascot. Entry names are according to SwissProt and Uniprot databases. Candidates were detected in at least two from four independent experiments. Table S3. Silica NP-specific C. elegans aggregome as identified by mass spectrometry analysis. Entry name Protein names translation, rRNA metabolic process EF2_CAEEL Elongation factor 2 (EF-2) FBRL_CAEEL rRNA 2'-O-methyltransferase fibrillarin (EC 2.1.1.-) (Histone-glutamine methyltransferase) GLH1_CAEEL ATP-dependent RNA helicase glh-1 (EC 3.6.4.13) (Germline helicase 1) IF4A_CAEEL IF5A1_CAEEL Eukaryotic initiation factor 4A (eIF-4A) (EC 3.6.4.13) (ATP-dependent RNA helicase eIF4A) (Initiation factor 1) Eukaryotic translation initiation factor 5A-1 (eIF-5A-1) (Initiation factor five protein 1) RL11_CAEEL 60S ribosomal protein L11 RL13_CAEEL 60S ribosomal protein L13 RL18_CAEEL 60S ribosomal protein L18 RL21_CAEEL 60S ribosomal protein L21 RL22_CAEEL 60S ribosomal protein L22 RL36_CAEEL 60S ribosomal protein L36 RL7_CAEEL 60S ribosomal protein L7 RL8_CAEEL 60S ribosomal protein L8 RLA0_CAEEL 60S acidic ribosomal protein P0 RLA1_CAEEL 60S acidic ribosomal protein P1 (Ribosomal protein large subunit P1) RLA2_CAEEL 60S acidic ribosomal protein P2 RS12_CAEEL 40S ribosomal protein S12 RS16_CAEEL 40S ribosomal protein S16 RS17_CAEEL 40S ribosomal protein S17 RS19_CAEEL 40S ribosomal protein S19 RS27_CAEEL 40S ribosomal protein S27 RS28_CAEEL 40S ribosomal protein S28 RS4_CAEEL 40S ribosomal protein S4 RS5_CAEEL 40S ribosomal protein S5 RS6_CAEEL 40S ribosomal protein S6 SYDC_CAEEL Aspartate--tRNA ligase, cytoplasmic (EC 6.1.1.12) (Aspartyl-tRNA synthetase) (AspRS) YZVL_CAEEL Uncharacterized NOP5 family protein K07C5.4 protein folding, proteolysis, stress response ADF1_CAEEL Actin-depolymerizing factor 1, isoforms a/b (Uncoordinated protein 60) ASP6_CAEEL Aspartic protease 6 (EC 3.4.23.-) CH60_CAEEL Chaperonin homolog Hsp-60, mitochondrial (Heat shock protein 60) (HSP-60) CYP7_CAEEL Peptidyl-prolyl cis-trans isomerase 7 (PPIase 7) (EC 5.2.1.8) (Cyclophilin-7) (Rotamase 7) GPX2_CAEEL Glutathione peroxidase 2 (EC 1.11.1.9) HSP7C_CAEEL Heat shock 70 kDa protein C HSP7F_CAEEL Heat shock 70 kDa protein F, mitochondrial PSMD1_CAEEL 26S proteasome non-ATPase regulatory subunit 1 (26S proteasome regulatory subunit rpn-2) PSMD3_CAEEL 26S proteasome non-ATPase regulatory subunit 3 (26S proteasome regulatory subunit rpn-3) TERA1_CAEEL NACA_CAEEL Transitional endoplasmic reticulum ATPase homolog 1 (Cell division cycle-related protein 48.1) (p97/CDC48 homolog 1) Nascent polypeptide-associated complex subunit alpha (NAC-alpha) (Alpha-NAC) muscle contraction DIM_CAEEL Disorganized muscle protein 1 (2D-page protein spot 8) MLE_CAEEL Myosin, essential light chain (Myosin light chain alkali) TPM1_CAEEL Tropomyosin isoforms a/b/d/f (Levamisole resistant protein 11) UNC87_CAEEL Protein unc-87 (Uncoordinated protein 87) metabolic processes ACBP1_CAEEL Acyl-CoA-binding protein homolog 1 (ACBP-1) (Diazepam-binding inhibitor homolog) (DBI) ACON_CAEEL Probable aconitate hydratase, mitochondrial (Aconitase) (EC 4.2.1.3) (Citrate hydro-lyase) ALF2_CAEEL Fructose-bisphosphate aldolase 2 (EC 4.1.2.13) (Aldolase CE-2) (CE2) BCAT_CAEEL Branched-chain-amino-acid aminotransferase, cytosolic (BCAT) (EC 2.6.1.42) (ECA39 protein) DHSA_CAEEL DLDH_CAEEL Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial (EC 1.3.5.1) (Flavoprotein subunit of complex II) (FP) Dihydrolipoyl dehydrogenase, mitochondrial (EC 1.8.1.4) (Dihydrolipoamide dehydrogenase) G3P2_CAEEL Glyceraldehyde-3-phosphate dehydrogenase 2 (GAPDH-2) (EC 1.2.1.12) G3P3_CAEEL Glyceraldehyde-3-phosphate dehydrogenase 3 (GAPDH-3) (EC 1.2.1.12) Q23621_CAEE L GLYC_CAEEL Glutamate dehydrogenase HACD_CAEEL IDH3A_CAEEL MCCB_CAEEL MDHM_CAEEL PCCA_CAEEL SAHH_CAEEL TECR_CAEEL Serine hydroxymethyltransferase (SHMT) (EC 2.1.2.1) (Glycine hydroxymethyltransferase) (Glycosylationrelated protein 1) (Maternal effect lethal protein 32) (Serine methylase) Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase hpo-8 (EC 4.2.1.134) (Probable 3-hydroxyacyl-CoA dehydratase) Probable isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial (EC 1.1.1.41) (Isocitric dehydrogenase subunit alpha) (NAD(+)-specific ICDH subunit alpha) Probable methylcrotonoyl-CoA carboxylase beta chain, mitochondrial (MCCase subunit beta) (EC 6.4.1.4) (3-methylcrotonyl-CoA carboxylase 2) (3-methylcrotonyl-CoA carboxylase non-biotin-containing subunit) (3methylcrotonyl-CoA:carbon dioxide ligase subunit beta) Probable malate dehydrogenase, mitochondrial (EC 1.1.1.37) Propionyl-CoA carboxylase alpha chain, mitochondrial (PCCase subunit alpha) (EC 6.4.1.3) (PropanoylCoA:carbon dioxide ligase subunit alpha) Adenosylhomocysteinase (AdoHcyase) (EC 3.3.1.1) (Protein dumpy-14) (S-adenosyl-L-homocysteine hydrolase) Probable very-long-chain enoyl-CoA reductase art-1 (EC 1.3.1.93) ion transport G5EFU2_CAEEL Mitochondrial adenine nucleotide translocase 1.4 (Protein ANT-1.4) G5EFW8_CAEEL Mitochondrial adenine nucleotide translocase 1.3 (Protein ANT-1.3) VATH2_CAEEL Probable V-type proton ATPase subunit H 2 (V-ATPase subunit H 2) (Vacuolar proton pump subunit H 2) cellular structure H2B1_CAEEL Histone H2B 1 IFC2_CAEEL Intermediate filament protein ifc-2 (Cel IF C2) (Intermediate filament protein C2) (IF-C2) TCTP_CAEEL Translationally-controlled tumor protein homolog (TCTP) transport PGP1_CAEEL Multidrug resistance protein pgp-1 (EC 3.6.3.44) (P-glycoprotein A) (P-glycoprotein-related protein 1) Q94148_CAEEL Protein RAB-10 others LEC1_CAEEL 32 kDa beta-galactoside-binding lectin (32 kDa GBP), lec-1 F37C4_CAEEL Protein F37C4.5 Table S3 lists all filter-trapped SDS-insoluble proteins specifically detected in silica NPtreated 2-day-old, adult wild-type C. elegans. Candidates were characterized by mass spectrometry and measured spectra were searched against the SwissProt database by Mascot. Entry names are according to SwissProt and Uniprot databases. Candidates were detected in at least two from four independent experiments. Video S1. The movie visualizes the locomotion pattern of a representative 2-day-old (young adult) wild type C. elegans nematode that was mock (H2O)-treated for 24 hours. The video is slowed down to 81% real-time with 15 frames / second. The worm crawls in regular sinusoidal curves. Video S2. The movie visualizes the locomotion pattern of a representative 2-day-old (young adult) wild type C. elegans nematode that was exposed to silica NPs for 24 hours. The video is slowed down to 81% real-time with 15 frames / second. The worm crawls in a serrated locomotion pattern and covers a shorter distance compared to the untreated worm (Video S1). Video S3. The movie visualizes the locomotion pattern of a representative 2-day-old (young adult) wild type C. elegans nematode that was exposed to BULK silica for 24 hours. The video is slowed down to 81% real-time with 15 frames / second. The worm crawls in regular sinusoidal curves that are comparable to the forward locomotion of untreated worms (Video S1). The speed is likewise similar to the one observed in untreated worms.