Sample Size Determination
advertisement

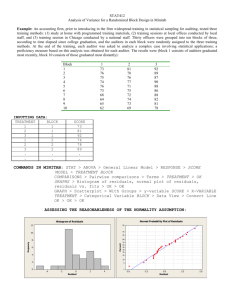
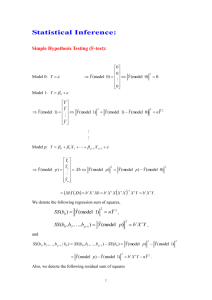
ST 524 NCSU - Fall 2008 RCBD and Power Types of Error in Hypothesis testing To compare two treatment means, we carry out a test of We make a Type I error if H o : 1 2 against H1 : 1 2 . H o is true 1 2 but we conclude H1 1 2 . H o when in fact 1 2 . We make a Type II error if we fail to reject Power = P(Type I Error) = probability of a Type II error, Power = probability that the test will reject H o , given that H o is false. True Situation Decision Taken 1 2 1 2 Do Not Reject Ho Reject Ho Correct Decision Type I Error Type II Error Correct Decision =P Reject Ho/1 2 P Type I Error =P Do not Reject Ho/1 2 P Type II Error Power P Reject Ho/1 2 1 Power For testing H o : 1 2 against H1 : 1 2 , power depends on: a) True value of 1 2 , b) , measure of variability. c) r, number of repetitions per treatment, and on d) , significance level of test. 2 Power increases as r 1 2 2 2 increases. Sample size Sample Size Determination For a Randomized Complete Block Design Yij i b j eij iidN 0, 2 eij i is the effect of ith treatment i i ' difference between means for treatments i and i’. Number of repetitions per treatment is equal to the number of blocks in a balanced design. Number of replicates required depends on Hypothesis is being tested Whether test is one- or two-tailed (what are the possible alternative hypotheses?) Significance level, , to be used, P(Type I Error). 2 1 Size of the difference What assurance is desired to detect the difference ( Power 1 )? An estimate of the variability of the data. 2 . to be detected. Sample size required to detect a mean difference at least equal to Tuesday September 4, 2008 1 2 o is given by 1 ST 524 NCSU - Fall 2008 RCBD and Power r 2 2 z 2 z 2 Express the desired difference to detect, , as a multiple of the true standard deviation r 2 z 2 z 2 2 2 Correction when residual variance is used instead of true variance Error df 3 rnew r Error df 1 Example 1 Table 9.2 ST&D (p. 207) Oil Content of Redwing Flaxseed inoculated at different stages of growth with S. linicola, Winnipeg, 1947 (in percentage) Analysis of Variance Source of Variation Blocks Treatments Error Total df r-1 = 3 t–1=5 (r-1)(t-1) = 15 n – 1= 23 SS MS F 3.14 31.65 19.72 54.51 1.05 6.33 1.31 4.83 Calculate the number of repetitions per treatment to detect a difference effect between treatments of at least 2.5% oil, regardless of direction, at a significance level of 0.05 with a 90% assurance of detecting a true difference of 2.5% 2 1.311.96 1.28 4.4 , 2.52 2 r We need r = 5 blocks to attain desired power in detecting a difference effect of 2.5% Example 2. Calculate sample size for detecting difference D between clones 2 and 5, MSE = 11793 D n 55 82 75 45 95 28 115 19 135 14 165 10 185 8 205 6 225 5 255 4 80 n=2*(11793)*(1.96+1.28)^2/(c(55,75,95,115,135,165,185,205,225,255)^2) n [1] 81.850047 44.017137 27.434503 18.721845 13.585536 9.094450 7.234372 [8] 5.891645 4.890793 3.807711 n 20 40 60 power=0.90 var=11793 50 100 150 200 250 D Tuesday September 4, 2008 2 ST 524 NCSU - Fall 2008 RCBD and Power Randomized Block Design treat block_1 block_2 block_3 block_4 Early_Bloom 33.3 31.9 34.9 37.1 Full_Bloom 34.4 34.0 34.5 33.1 Full_Bloom_P 36.8 36.6 37.0 36.4 Ripening 36.3 34.9 35.9 37.1 Seedling 34.4 35.9 36.0 34.1 Uninoculated 36.4 37.3 37.7 36.7 Field Layout block plot_1 plot_2 plot_3 plot_4 plot_5 plot_6 1 6 2 3 5 4 1 2 5 4 3 2 1 6 3 3 1 6 5 4 2 4 4 2 5 6 1 3 Linear Model Treatments and Block as fixed effect factors Yij i j eij 33.3 1 31.9 1 34.9 1 37.1 1 34.4 1 34.0 1 34.5 1 33.1 1 36.8 1 36.6 1 37.0 1 36.4 1 36.3 1 34.9 1 35.9 1 37.1 1 34.4 1 35.9 1 36.0 1 34.1 1 36.4 1 37.3 1 37.7 1 36.7 1 1 0 0 0 0 0 1 0 0 0 e11 e 1 0 0 12 e13 0 0 1 0 0 0 0 1 e14 e21 1 0 0 0 0 1 0 0 e22 e 0 0 1 0 23 0 0 0 1 e24 1 1 0 0 0 e31 2 0 1 0 0 e32 3 0 0 1 0 e33 4 0 0 0 1 e34 5 1 0 0 0 e41 6 0 1 0 0 e42 1 0 0 1 0 e43 2 0 0 0 1 e44 3 1 0 0 0 e51 4 0 1 0 0 e52 e53 0 0 1 0 0 0 0 1 e54 e61 1 0 0 0 0 1 0 0 e62 e 0 0 1 0 63 0 0 0 1 e64 1 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 1 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 1 1 Y Xβ e , eij ~ iidN 0, e2 Tuesday September 4, 2008 title "RCBD Block and Treat fixed effects"; proc mixed data=redwing; class block treat; model y= block treat ; run; 3 ST 524 NCSU - Fall 2008 RCBD and Power Model Information Data Set Dependent Variable Covariance Structure Estimation Method Residual Variance Method Fixed Effects SE Method Degrees of Freedom Method WORK.REDWING y Diagonal REML Profile Model-Based Residual Class Level Information Class Levels block treat 4 6 Values 1 2 3 4 Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated Dimensions Covariance Parameters Columns in X Columns in Z Subjects Max Obs Per Subject 1 11 0 1 24 Number of Observations Number of Observations Read Number of Observations Used Number of Observations Not Used 24 24 0 The Mixed Procedure Covariance Parameter Estimates Cov Parm Estimate Residual 1.3144 ˆ e2 1.3144 Fit Statistics -2 Res Log Likelihood AIC (smaller is better) AICC (smaller is better) BIC (smaller is better) 59.0 61.0 61.3 61.7 Type 3 Tests of Fixed Effects Effect block treat Num DF Den DF F Value Pr > F 3 5 15 15 0.80 4.82 0.5147 0.0080 Block and Treat are fixed-effect factors Expected Mean Squares ANALYSIS of VARIANCE TABLE Type 3 Analysis of Variance Source DF Sum of Squares block treat Residual 3 5 15 3.141250 31.652083 19.716250 Mean Square 1.047083 6.330417 1.314417 Tuesday September 4, 2008 Expected Mean Square Error Term Var(Residual) + Q(block) Var(Residual) + Q(treat) Var(Residual) MS(Residual) MS(Residual) . Error DF F Value Pr > F 15 15 . 0.80 4.82 . 0.5147 0.0080 . 4 ST 524 NCSU - Fall 2008 RCBD and Power r i t Q Treat i 1 Var(Residual) t 1 is t j r 2 Q Block , j 1 2 r 1 2 e Test of Hypothesis a. Block H o : 1 2 3 4 0 H1 : at least one j 0, j 1 4 b. Treatments H o : 1 2 3 4 5 6 0 H1 : at least one i 0, p value= 0.0080, Reject Ho at 0.05 significance level. i 1 6 Treatments as fixed-effect factor and Block as random-effect factor Yij i b j eij b j ~ iidN 0, b2 , eij ~ iidN 0, e2 33.3 1 31.9 1 34.9 1 37.1 1 34.4 1 34.0 1 34.5 1 33.1 1 36.8 1 36.6 1 37.0 1 36.4 1 36.3 1 34.9 1 35.9 1 37.1 1 34.4 1 35.9 1 36.0 1 34.1 1 36.4 1 37.3 1 37.7 1 36.7 1 1 0 0 0 0 0 1 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 0 1 0 0 0 0 0 0 1 0 0 0 0 1 0 0 1 0 0 0 0 0 0 0 1 0 0 0 1 0 0 1 0 0 0 2 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 3 1 0 0 0 1 0 0 0 0 0 0 1 0 0 0 0 0 0 1 0 0 4 0 0 0 0 0 1 0 1 0 0 0 0 1 0 0 0 0 0 0 1 0 5 0 0 0 0 0 1 0 0 0 0 0 0 0 1 6 1 0 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 1 0 Y Xβ Zb e 0 0 0 e11 e 1 0 0 12 e13 0 1 0 0 0 1 e14 e21 0 0 0 1 0 0 e22 e 0 1 0 23 0 0 1 e24 e 0 0 0 31 e32 1 0 0 0 1 0 b1 e33 0 0 1 b2 e34 0 0 0 b3 e41 1 0 0 b4 e42 0 1 0 e43 e 0 0 1 44 0 0 0 e51 1 0 0 e52 e53 0 1 0 0 0 1 e56 e61 0 0 0 1 0 0 e62 e 0 1 0 63 0 0 1 e64 title "RCBD Block and Treat fixed effects"; proc mixed data=redwing; class block treat; model y= treat ; random intercept / subject=block; run; Model Information Data Set Dependent Variable Covariance Structure Subject Effect Estimation Method Residual Variance Method Fixed Effects SE Method Degrees of Freedom Method WORK.REDWING y Variance Components block REML Profile Model-Based Satterthwaite Class Level Information Tuesday September 4, 2008 5 ST 524 NCSU - Fall 2008 RCBD and Power Class block treat Levels 4 6 Values 1 2 3 4 Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated Dimensions Covariance Parameters Columns in X Columns in Z Per Subject Subjects Max Obs Per Subject 2 7 1 4 6 Number of Observations Number of Observations Read Number of Observations Used Number of Observations Not Used 24 24 0 Covariance Parameter Estimates Cov Parm Estimate block Residual 0 1.2699 > 3.141250+19.716250 [1] 22.8575 > 22.8575/18 [1] 1.269861 Fit Statistics -2 Res Log Likelihood AIC (smaller is better) AICC (smaller is better) BIC (smaller is better) 63.7 65.7 65.9 65.1 Type 3 Tests of Fixed Effects Effect treat Num DF Den DF F Value Pr > F 5 15 4.99 0.0069 > 6.330417/1.2699 [1] 4.984973 How many degrees of freedom for Error? 15+3 = 18 Block is random-effect factor and Treat is fixed-effect factor Type 3 Analysis of Variance Source DF Sum of Squares treat block Residual 5 3 15 31.652083 3.141250 19.716250 Var(block) is b2 Var(Residual) is Mean Square 6.330417 1.047083 1.314417 Error Term Var(Residual) + Q(treat) Var(Residual) + 6 Var(block) Var(Residual) MS(Residual) MS(Residual) . Error DF F Value Pr > F 15 15 . 4.82 0.80 . 0.0080 0.5147 . e2 Method of Moments to estimate ˆ b2 Expected Mean Square b2 BlockMS ErrorMS 1.047083 1.314417 -0.04456 t 6 Since estimated value for b2 is negative we can assume that variance for block effects is 0. Tuesday September 4, 2008 6 ST 524 NCSU - Fall 2008 RCBD and Power Need to correct the number of degrees of freedom in Type 3 test of hypothesis for fixed effects RCBD Block random effects and Treat fixed effects Satterthwaite correction for degrees of freedom Covariance Parameter Estimates title "RCBD Block and Treat fixed effects"; proc mixed data=redwing; class block treat; model y= treat/ddfm=satter ; random intercept / subject=block; run; Cov Parm Estimate block Residual 0 1.2699 ˆ b2 0 ˆ e2 1.2699 Fit Statistics -2 Res Log Likelihood AIC (smaller is better) AICC (smaller is better) BIC (smaller is better) 63.7 65.7 65.9 65.1 Type 3 Tests of Fixed Effects Effect Num DF Den DF F Value Pr > F 5 18 4.99 0.0049 treat Test of Hypothesis 2 b. Block H o : b 0 c. H1 : b2 0 Treatments H o : 1 2 3 4 5 6 0 H1 : at least one i 0, i 1 6 p value= 0.0049, Reject Ho at 0.05 significance level. Power Study Assume a RCBD, Fixed Effects, with t treatments and r blocks. Assume that the true treatment means are 1 , 2 , * , 6* , and that the error variance is e2 . * The power of the test to test H o : 1 2 t H1 : at least one i is different from others, i 1 t depends on the noncentrality t parameter , where 1 e2 r t * i and * 2 i * i 1 * i t . As the noncentrality parameter increases, power increases. To compute power for a given number of reps ® and a given alternative H1, that specifies values for 1* , , t* we may use SAS: b. c. 1* , , t* under H1 and compute . State significance level and critical value for testing null hypothesis State e2 and values for a. H o : 1 2 5 t , From ANOVA table for RCBD, the test statistic is F TreatmentMS , with (t-1) and (t-1)(r-1) ErrorMS degrees of freedom. b. Critical value of F distribution is c. Calculate power as Fcrit Ft 1,t 1 r 1, = F Hdf , Edf Power P Reject H o | H1 P Fcalc Fcrit | , Hdf , Edf 1 P Fcalc Fcrit | , Hdf , Edf Tuesday September 4, 2008 7 ST 524 NCSU - Fall 2008 RCBD and Power Example, based in results for example 9.2 (STD) Error MS = 1.3144 Treatment means: 34.3, 34, 36.7, 36.05, 35.1, 37.025 and Overall mean = 35.5292, = 0.05, Fcrit = F(5,15,0.05) = 2.9013 Obs grndmn dfnum mse t r trt mu 1 2 3 4 5 6 35.5292 35.5292 35.5292 35.5292 35.5292 35.5292 5 5 5 5 5 5 1.31 1.31 1.31 1.31 1.31 1.31 6 6 6 6 6 6 4 4 4 4 4 4 A B C D E F 34.300 34.000 36.700 36.050 35.100 37.025 Noncentrality parameter == 24.0810 Power = 1- P(F<2.9013| 24.0810,5,15) = diff_mn2 1.51085 2.33835 1.37085 0.27127 0.18418 2.23752 term 4.61328 7.14000 4.18580 0.82830 0.56239 6.83211 0.90594 Power vs sample size - example STD-9.2 How does power change as number of blocks (repetitions) varies? Power vs lambda (noncentrality parameter) - example STD-9.2 How does power change asnoncentrality parameter varies? Tuesday September 4, 2008 8