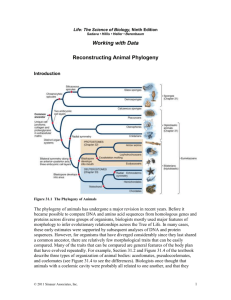
Mammalian Phylogenetic Systematics

Rev 1/15
Molecular Phylogenetics Exercise
Name ____________________________
Walruses and whales and seals, oh my!
Walruses and whales are both marine eutherians (placental mammals). So are dolphins, seals, and manatee. They all have streamlined bodies, legs reduced to flippers, blubber under the skin and other adaptations for survival in the water. Although mammals evolved on land, these species have returned to the sea. Did they evolve from a single ancestor who returned to the ocean, or were there different return events and parallel evolution? We can’t go back in time to observe what happened, but DNA sequences contain evidence about the relationships of living creatures.
From these relationships, we can learn about the evolutionary history of marine mammals.
In this exercise, we will use DNA sequence information in
GenBank (the public repository of all known DNA sequences from many species) and phylogenetics software to test hypotheses about the relationship between aquatic mammals (seals, whales, dolphins, walruses, manatees) and their potential ancestral relationship to land mammals. We will use a protein that all mammals share: the hemoglobin beta protein. Hemoglobin is a good test molecule since it shows both conservation across species (since it performs the essential function of carrying oxygen in the blood), and variation between species. Species with unique challenges such as holding their breath for long underwater dives, may have evolved changes in their hemoglobin which improved their supply of oxygen. In addition, hemoglobin has been studied by many evolutionary biologists, so sequences are available in GenBank from many different organisms.
Procedure
In this exercise, we will be testing hypotheses about the evolutionary ancestry of different marine mammals. To repeat, we are trying to answer the question: Did marine mammals evolve from a single ancestor who returned to the ocean, or were there distinct return events from separate ancestors? As a starting point let’s hypothesize that marine mammals have a single common land mammal ancestor.
First, we will explore the relationship of the marine mammals to each other vs. their evolutionary relationship to other placental mammals. To do this, we will test whether seals and whales are more closely related to each other than either of them are to terrestrial and winged representatives of major eutherian groups. You will also use the phylogeny to test whether modern eutherian taxonomy accurately reflects the evolutionary relationships among different placental mammals.
Homework
Complete steps 1–8 prior to our laboratory meeting
Finding amino acid sequences
1. First we need to get the sequence (amino acid) data for the hemoglobin protein from our marine and land animals listed in the Appendix 1 (both the ingroup & outgroup taxa). Go to GenBank , a DNA and protein sequence database hosted by the National Center for Biotechnology Information (NCBI) in Maryland at: http://www.ncbi.nlm.nih.gov/Genbank/ (Note: You can also go to Google and search for “GenBank,” but it is not always the first website that pops up.)
2. One thing you need to know, is that the hemoglobin beta gene and protein is known as “ HBB ” in GenBank. In the “Search” window, select “Protein” from the pull-down menu, because we want to compare the amino acid sequence from each animal for this protein. We need to be specific about the identity of each animal, so use the name of the organism you are searching for along with HBB as search terms. Select “Go”.
NOTE : This database was developed by humans, so sometimes it is not organized logically. Don’t get frustrated; be patient. If your search returns several answers that you have to choose between, look for the listing with a “P0####” number and a length of 146-147 aa. That will be the correct protein!
3. The search result is a page with a lot of information about the protein from this organism. To see the actual amino sequence for this protein, click on the “FASTA” link near the top of the page.
Page 2
4. The FASTA page presents the amino acid sequence of the protein in a coded format using single letters to represent each of the 20 amino acids (A=alanine, M=methionine, P=proline, etc.; Appendix 2 ). Copy the amino acid sequence. You must include the header line, starting from the greater than symbol (>). THIS IS
IMPORTANT! Make sure the sequence of amino acids is 146-147 aa long.
Creating a Fasta File
5. Paste the amino acid sequence into a text file on your computer. Open up NotePad (on a PC) or Microsoft
Word (on a Mac). Save as type “All Files” from the dropdown and name it “Eutheria_Phylogeny.fas”. We will refer to this as your “FASTA text file”.
6. Go back to GenBank and collect the amino acid sequences for the HBB gene from each species and paste them onto separate lines in the same FASTA text file. Adjust the header for each sequence as depicted below
(i.e. >Sloth rather than >Pale_Throated_Sloth and especially not >Bradypus_tridactylus).
Tip: If you want to use more than one word in your label, like “harbor seal” you must add an underscore “_” between the words (harbor_seal) instead of a space between words. This is the only way that all the words will show up as labels on your tree. Do not use names longer than 30 characters.
Page 3
7. Scan through your FASTA text file, it is critically important that it is formatted correctly. There must be a
“paragraph return” or “hard return” (created by the Enter key) only after your header and only after the complete end of the sequence. Although it may appear that a hard return is already there, it is good practice to add one, because the hidden characters do not always cut and paste correctly. Check again to be sure each sequence is the same length as the others (146-147 aa). Save your work.
For example:
8. Download MEGA 6 from http://megasoftware.net/. This program, which stands for Molecular Evolutionary
Genetic Analysis, is free and will be used to align your sequences, calculate genetic distances, and to construct a phylogeny. Having the version on your own computer will allow you to check your lab work at home!
STOP HOMEWORK HERE!
The remaining sections will be completed in laboratory
Aligning Sequences
9. Open MEGA 6 and choose Edit/Build Alignment from the Align dropdown menu. Select Retrieve sequences from a file in the popup window and click OK . Select the file that you created and click Open .
Notice that the amino acids are color coded, and that the colors are not lined up. We need to fix this by performing a sequence ‘alignment’. Note: These steps work on a small internal window, not from the top menu bar.
10. Highlight your data set by selecting all of the taxon names on the left (select the first and last taxa while holding the shift key). In the Alignment menu choose Align by Muscle . Alignments can often be tricky so there are lots of parameters that can be adjusted in the window that pops up. This is a relatively simple data set, however, so the default parameters will work just fine. Click Compute . Click on a column or row header to deselect the taxa.
Your sequences should now be nicely aligned.
11. You need to save your alignment, so from the Data menu select Export Alignment and then MEGA format from the Data menu. Save your file as “Eutheria_Alignment”.
Is this starting to look familiar to you? It should! You just constructed a data matrix, not that different from the matrices you analyzed for the Clade Critters and Cyanobacteria exercises.
This amino acid matrix differs from the morphological matrix in the Clade Critters exercise by having
__________ rather than __________ possible character states.
This amino acid matrix differs from the nucleotide matrix in the Cyanobacteria exercise by having
__________ rather than __________ possible character states.
12. Close the alignment file (but not MEGA).
/4
Page 4
Analyzing the Data
13. Now that you have an alignment you can conduct some interesting analyses. First, let’s explore genetic distances between the different taxa. Open your saved alignment in MEGA. From the Distances menu select Compute Pairwise Distances and click YES in the pop up window. Another window will display.
Accept the default settings except one, change Model/Method to p-distance . This is a simple metric that represents the proportion (p) of amino acid sites at which two sequences being compared are different. It is obtained by dividing the number of amino acid differences by the total number of amino acids being compared. For instance, if you compared two amino acid sequences, each 100 amino acids long, and they differed at 10 positions, then the p-distance for this pairwise comparison would be 0.10 (10%). Click
Compute and open the resulting pairwise distance matrix.
What is the p-distance between the following taxa
Harbor Seal & Sperm Whale = ______________
Harbor Seal & Minke Whale = ______________
Rhinoceros & Horse = ______________
Horse & Elephant = ______________
Sperm Whale & Minke Whale = ______________ Human & Rhesus Macaque = _____________
14. Close the distance matrix and from the phylogeny menu select Construct/Test Neighbor-Joining Tree .
Use the currently active data. A window will appear with parameters that you can set before constructing a neighbor-joining (NJ) phylogeny. Change “Test of Phylogeny” to Bootstrap Method and change the “No. of Bootstrap Replications” to 1000 . Make sure that the “Model/Method” is set to p-distance and that
“Gaps/Missing Data Treatment” is set to Complete deletion .
15. Click Compute . The analysis will run for a moment (NJ is fast!) and MEGA will then display a phylogeny.
Your instructor will explain how NJ phylogenies are constructed and how to interpret the bootstrap values at each node. Display this Traditional Tree , including bootstrap values, and save the phylogeny as a PDF by selecting the Image dropdown menu and selecting Save as PDF file.
16. To explore another method of visualizing a NJ phylogeny, from the View menu select Tree/Branch style and then click Radiation. Display this Radiation Tree , including bootstrap values, and Save the Radiation
Tree as a PDF.
17. Print out both phylogenies and attach them to this handout. (worth 5 points each)
/16
Page 5
Congratulations, you just conducted your first phylogenetic analysis!
Now answer the following questions based on your results.
We conducted this exercise to address whether ______________ ______________ evolved from a single ancestor that returned to the ocean.
To do this, we tested whether ______________ and ______________ are more closely related to each other than either of them are to terrestrial and winged representatives of major eutherian groups.
Based on our analysis, I reject fail to reject this hypothesis .
The phylogenetic analysis of HBB sequences suggested that eutherians returned to aquatic and semi-aquatic habitats a minimum of ______ times.
Bootstrap values provide the strongest support for a sister relationship between _____________________ and _____________________.
Draw lines connecting taxa with their closest relatives, based on results from your analysis.
Homo sapiens Hippopotamus amphibius
Bos taurus
Physeter catodon
Canis lupus familiaris
Tursiops truncates
Ursus maritimus
Macaca mulatta
Based on our analysis, Carnivora is monophyletic paraphyletic polyphyletic .
Based on our analysis, Artiodactyla is monophyletic paraphyletic polyphyletic .
Based on our analysis, Cetacea is monophyletic paraphyletic polyphyletic .
Based on our analysis, marine mammals are monophyletic paraphyletic polyphyletic .
What does the biologist need to do to understand the basal (weakly supported) relationships in this phylogeny? (2pt)
__________________________________________________________________________
__________________________________________________________________________
/17
Page 6
APPENDIX 1
Class Mammalia
Eutheria Clade
Order Pilosa
Bradypus tridactylus (Pale-throated Sloth)
Order Cingulata
Dasypus novemcinctus (Nine-banded Armadillo)
Order Lagomorpha
Lepus europaeus (European Hare)
Order Rodentia
Mus musculus (House Mouse)
Rattus norvegicus
Order Primates
(Brown Rat)
Macaca mulatta (Rhesus Macaque)
Homo sapiens (Humans)
Order Chiroptera
Antrozous pallidus (Pallid Bat)
Rousettus aegyptiacus (Egyptian Fruit Bat)
Order Insectivora
Condylura cristata (Star-nosed Mole)
Order Carnivora
Canis lupus familiaris (Domestic Dog)
Ursus maritimus (Polar Bear)
Phoca vitulina (Harbor Seal)
Odobenus rosmarus divergens (Walrus)
Order Artiodactyla
Bos taurus (Cow)
Camelus bactrianus (Bactrian Camel)
Hippopotamus amphibius (Hippopotamus)
Order Cetacea
Physeter catodon (Sperm Whale)
Tursiops truncates (Bottlenosed Dolphin)
Balaenoptera acutorostrata (Minke Whale)
Order Perissodactyla
Equus caballus (Horse)
Rhinoceros unicornis (Indian Rhinoceros)
Order Sirenia
Trichechus inunguis (Amazonian Manatee)
Order Proboscidae
Loxodonta Africana (African Elephant)
Marsupalia Clade
Order Diprotodontia
Macropus giganteus (Eastern Grey Kangaroo)
Order Didelphimorphia
Didelphis virginiana (Virginia Opossum)
Monotreme Clade
Order Monotremata
Ornithorhynchus anatinus (Platypus)
INGROUP
OUTGROUP
Page 7
APPENDIX 2
Page 8