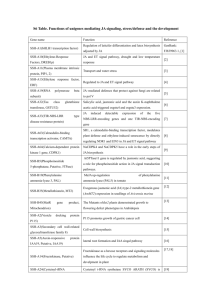
Supplemental Table 5 Locus information and gene names
advertisement
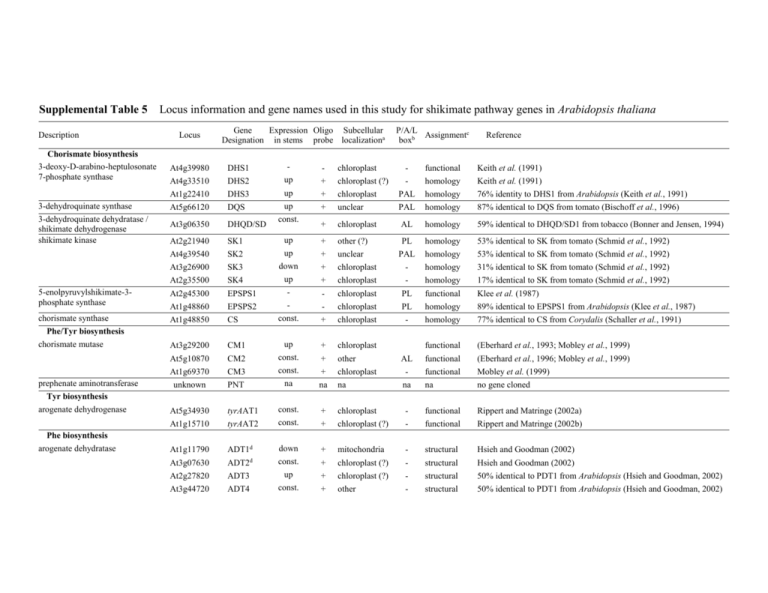
Supplemental Table 5 Description Locus information and gene names used in this study for shikimate pathway genes in Arabidopsis thaliana Locus Gene Expression Oligo Subcellular Designation in stems probe localizationa Chorismate biosynthesis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase At4g39980 At4g33510 DHS1 DHS2 up + chloroplast chloroplast (?) 3-dehydroquinate synthase At1g22410 At5g66120 DHS3 DQS up up + + At3g06350 DHQD/SD At2g21940 At4g39540 SK1 SK2 At3g26900 At2g35500 SK3 SK4 5-enolpyruvylshikimate-3phosphate synthase At2g45300 At1g48860 EPSPS1 EPSPS2 chorismate synthase Phe/Tyr biosynthesis chorismate mutase At1g48850 CS At3g29200 3-dehydroquinate dehydratase / shikimate dehydrogenase shikimate kinase prephenate aminotransferase Tyr biosynthesis arogenate dehydrogenase Phe biosynthesis arogenate dehydratase P/A/L Assignmentc boxb Reference - functional homology Keith et al. (1991) Keith et al. (1991) chloroplast unclear PAL PAL homology homology 76% identity to DHS1 from Arabidopsis (Keith et al., 1991) 87% identical to DQS from tomato (Bischoff et al., 1996) + chloroplast AL homology 59% identical to DHQD/SD1 from tobacco (Bonner and Jensen, 1994) up up + + other (?) unclear PL PAL homology homology 53% identical to SK from tomato (Schmid et al., 1992) 53% identical to SK from tomato (Schmid et al., 1992) down up + + chloroplast chloroplast - homology homology 31% identical to SK from tomato (Schmid et al., 1992) 17% identical to SK from tomato (Schmid et al., 1992) - - chloroplast chloroplast PL PL functional homology Klee et al. (1987) 89% identical to EPSPS1 from Arabidopsis (Klee et al., 1987) const. + chloroplast - homology 77% identical to CS from Corydalis (Schaller et al., 1991) CM1 up + chloroplast functional (Eberhard et al., 1993; Mobley et al., 1999) At5g10870 At1g69370 CM2 CM3 const. const. + + other chloroplast AL - functional functional (Eberhard et al., 1996; Mobley et al., 1999) Mobley et al. (1999) unknown PNT na na na na na no gene cloned const. const. + + chloroplast chloroplast (?) - functional functional Rippert and Matringe (2002a) Rippert and Matringe (2002b) const. At5g34930 At1g15710 tyrAAT1 tyrAAT2 At1g11790 ADT1d down + mitochondria - structural Hsieh and Goodman (2002) At3g07630 At2g27820 ADT2d ADT3 const. up + + chloroplast (?) chloroplast (?) - structural structural Hsieh and Goodman (2002) 50% identical to PDT1 from Arabidopsis (Hsieh and Goodman, 2002) At3g44720 ADT4 const. + other - structural 50% identical to PDT1 from Arabidopsis (Hsieh and Goodman, 2002) At5g22630 ADT5 up + chloroplast - structural 48% identical to PDT1 from Arabidopsis (Hsieh and Goodman, 2002) At1g08250 ADT6 up + chloroplast - structural 45% identical to PDT1 from Arabidopsis (Hsieh and Goodman, 2002) At5g05730 ASA1 const. + chloroplast (?) - functional Niyogi and Fink (1992) At2g29690 At3g55870 ASA2 ASA3 const. const. + + chloroplast other (?) - functional homology Niyogi and Fink (1992) 72% identical to ASA1 from Arabidopsis (Niyogi and Fink, 1992) At1g25220 At1g24807 ASB1 ASB2 const. const. + - chloroplast other AL - functional homology Niyogi et al. (1993) 97% identical to ASB1 from Arabidopsis (Niyogi et al., 1993) At1g24909 At1g25083 ASB3 ASB4 const. const. - other other - homology homology 97% identical to ASB1 from Arabidopsis (Niyogi et al., 1993) 97% identical to ASB1 from Arabidopsis (Niyogi et al., 1993) At1g25155 At5g57890 ASB5 ASB6 const. const. - other unclear - homology homology 97% identical to ASB1 from Arabidopsis (Niyogi et al., 1993) 93% identical to ASB1 from Arabidopsis (Niyogi et al., 1993) phosphoribosylanthranilate transferase At5g17990 At1g70570 PAT1 PAT2 const. const. + + chloroplast chloroplast PL - functional homology Rose et al., (1992) 14% identical to PAT1 from Arabidopsis (Rose et al., 1992) phosphoribosylanthranilate isomerase At1g07780 At5g05590 PAI1 PAI2 const. const. + - chloroplast chloroplast PL functional functional Li et al. (1995b) Li et al. (1995b) At1g29410 At2g04400 PAI3 IGPS1 const. const. + + chloroplast unclear PAL functional functional Li et al. (1995b) Li et al. (1995a) tryptophan synthase alpha subunit At5g48220 At3g54640 IGPS2 TSA1 const. const. + + chloroplast (?) chloroplast PL homology functional 63% identical to IGPS1 from Arabidopsis (Li et al., 1995a) Radwanski et al., (1995) tryptophan synthase beta subunit At4g02610 At5g54810 TSA2 TSB1 const. const. + + other chloroplast PA homology functional 74% identical to TSA1 from Arabidopsis (Radwanski et al., 1995) Last et al., (1991) Trp biosynthesis anthranilate synthase alpha subunit anthranilate synthase beta subunit indole-3-glycerol phosphate synthase const. At4g27070 TSB2 + chloroplast homology Last et al., (1991) const. At5g28237 TSB3 - unclear AL homology 56% identical to TSB1 from Arabidopsis (Last et al., 1991) a subcellular localization is based on targetP prediction (Emanuelsson et al., 2000), "other" denotes a subcellular localization other than chloroplast, mitochondrium, or secretory pathway, "(?)" denotes a poor probability for the given localization (class 4 or class 5) b Presence of P, A, and L promoter elements (Logemann et al., 1995) was determined using MotifMapper (Wanke et al., 2004), presence of motifs is indicated only if at least two of the three motifs are present in the -1000 bp region of the corresponding genes c "functional" denotes a biochemical and/or gentical proof, "homology" denotes that the annotation is based on sequence similarity to characterized plant genes only, "structural" denotes an annotation based on domain structure and sequence similarity to bacterial and/or fungal genes only d designated PDT1 and PDT2 in Hsieh and Goodman (2002) as homologous to the yeast prephenate dehydratase References for Supplemental Table 4 Bischoff, M., Rosler, J., Raesecke, H.R., Gorlach, J., Amrhein, N. and Schmid, J. (1996) Cloning of a cDNA encoding a 3dehydroquinate synthase from a higher plant, and analysis of the organ-specific and elicitor-induced expression of the corresponding gene. Plant Mol. Biol. 31, 69-76. Bonner, C.A. and Jensen, R.A. (1994) Cloning of cDNA encoding the bifunctional dehydroquinase.shikimate dehydrogenase of aromatic-amino-acid biosynthesis in Nicotiana tabacum. Biochem J. 302, 11-14. Eberhard, J., Ehrler, T.T., Epple, P., Felix, G., Raesecke, H.-R., Amrhein, N. and Schmid, J. (1996) Cytosolic and plastidic chorismate mutase isozymes from Arabidopsis thaliana: molecular characterization and enzymatic properties. Plant J 10, 815821. Eberhard, J., Raesecke, H.-R., Schmid, J. and Amrhein, N. (1993) Cloning and expression in yeast of a higher plant chorismate mutase Molecular cloning, sequencing of the cDNA and characterization of the Arabidopsis thaliana enzyme expressed in yeast. FEBS Letters 334, 233-236. Emanuelsson, O., Nielsen, H., Brunak, S. and von Heijne, G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005-1016. Hsieh, M.-H. and Goodman, H.M. (2002) Molecular characterization of a novel gene family encoding ACT domain repeat proteins in Arabidopsis. Plant Physiol. 130, 1797-1806. Keith, B., Dong, X., Ausubel, F. and Fink, G. (1991) Differential induction of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase Genes in Arabidopsis thaliana by wounding and pathogenic attack. Proc. Natl. Acad. Sci. USA 88, 8821-8825. Klee, H.J., Muskpf, Y.M. and Gasser, C.S. (1987) Cloning of an Arabidopsis thaliana gene encoding 5-enolpyruvylshikimate-3phosphate synthase: sequence analysis and manipulation to obtain glyphosate-tolerant plants. Mol. Gen. Genet. 210, 437-442. Last, R.L., Bissinger, P.H., Mahoney, D.J., Radwanski, E.R. and Fink, G.R. (1991) Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase beta genes. Plant Cell 3, 345–358. Li, J., Chen, S., Zhu, L. and Last, R.L. (1995a) Isolation of cDNAs Encoding the Tryptophan Pathway Enzyme Indole-3-Glycerol Phosphate Synthase from Arabidopsis thaliana. Plant Physiol. 108, 877-878. Li, J., Zhao, J., Rose, A.B., Schmidt, R. and Last, R.L. (1995b) Arabidopsis phosphoribosylanthranilate isomerase: molecular genetic analysis of triplicate tryptophan pathway genes. Plant Cell 7, 447-461. Logemann, E., Parniske, M. and Hahlbrock, K. (1995) Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl. Acad. Sci. USA 92, 5905-5909. Mobley, E.M., Kunkel, B.N. and Keith, B. (1999) Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana. Gene 240, 115-123. Niyogi, K.K. and Fink, G.R. (1992) Two anthranilate synthase genes in Arabidopsis: defense-related regulation of the tryptophan pathway. Plant Cell 4, 721–733. Niyogi, K.K., Last, R.L., Fink, G.R. and Keith, B. (1993) Suppressors of trp1 fluorescence identify a new arabidopsis gene, TRP4, encoding the anthranilate synthase beta subunit. Plant Cell 5, 1011–1027. Radwanski, E.R., Zhao, J. and Last, R.L. (1995) Arabidopsis thaliana tryptophan synthase alpha: gene cloning, expression, and subunit interaction. Mol. Gen. Genet. 248, 657-667. Rippert, P. and Matringe, M. (2002a) Molecular and biochemical characterization of an Arabidopsis thaliana arogenate dehydrogenase with two highly similar and active protein domains. Plant Mol. Biol. 48, 361-368. Rippert, P. and Matringe, M. (2002b) Purification and kinetic analysis of the two recombinant arogenate dehydrogenase isoforms of Arabidopsis thaliana. Eur. J. Biochem. 269, 4753-4761. Rose, A.B., Casselman, A.L. and Last, R.L. (1992) A phosphoribosylanthranilate transferase gene is defective in blue fluorescent Arabidopsis thaliana tryptophane mutants. Plant Physiol. 100, 582-592. Schaller, A., Schmid, J., Leibinger, U. and Amrhein, N. (1991) Molecular cloning and analysis of a cDNA coding for chorismate synthase from the higher plant Corydalis sempervirens Pers. J. Biol. Chem. 266, 21434-21438. Schmid, J., Schaller, A., Leibinger, U., Boll, W. and Amrhein, N. (1992) The in-vitro synthesized tomato shikimate kinase precursor is enzymatically active and is imported and processed to the mature enzyme by chloroplasts. Plant J. 2, 375-383. Wanke, D., Stüber, K. and Berendzen, K.W. (2004) Motif Mapper v3.1.2. www.mpiz-koeln.mpg.de/coupland/coupland/mm3/html.