Genetics Study Guide
advertisement

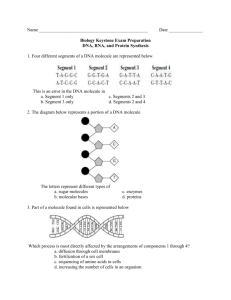
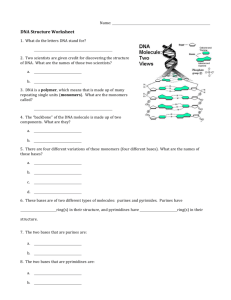
Genetics Study Guide- Chapter 2 1. Describe the composition of a nucleotide. 2. What are the characteristics of ribose and deoxyribose? 3. Which one are purines bases and which ones are pirimidine bases? How can you tell them apart? 4. How do the nitrogen bases attach to the pentose sugar? 5. What are triphosphate nucleotides? What is their function? 6. How are phosphodiester bonds form? 7. Are the first and the last nucleotide the same? 8. Explain the position of the second strand of the molecule of DNA. 9. How are the two strands linked together? 10. Explain Griffith experiments and define the transformation principle 11. What did Avery McLeod and McCarty found out. Describe their experiment and conclusions. 12. Describe Hershey &Chase experiment and conclusions. 13. What was Chargaff contribution to the discovery of the structure of molecule of DNA? 14. What was Franklin ’s contribution to the discovery of the structure of the molecule of DNA? 15. Explain the Watson and Crick model in detail. 16. How wide is the molecule of DNA? 17. What are the minor and major grooves? 18. What are the differences between A, B and Z DNA? 19. In addition to the nucleus, where in the cell can you find DNA? 20. Briefly describe the structure of a virus. What are bacteriophages? Based on their nucleic acids, what are the types of viruses that you can describe. 21. Describe the typical prokaryotic chromosome. How can the DNA of a bacteria fit inside the cell? What is the size of bacterial DNA? What is the function of topoisomerase enzymes? 22. What is a “looped domain?” Explain. 23. How do Eukaryotes organize their DNA? 24. Describe chromatin. What are the main types of proteins associated with the DNA. What is their electrical charged? Explain the formation of a nucleosome. 25. Once nucleosomes are formed, how is the chromatin organized? 26. What are the differences between euchromatin and heterochromatin and between constitutive heterochromatin and facultative heterochromatin. 27. What is centromeric and telomeric DNA. Adenine Histones Polymer Antiparallel Loop domains Purines Bacteriophages Major groove Pirimidines Chromatin Minor groove Ribonucleotide Chromosomes Monomers Ribonuclease Complementary base pairs Nitrogenous bases rRNA Constitutive heterochromatin Nucleic Acids Sort interspersed repeated sequences (SINEs) Nucleoid Cytosine Tandemly repeated DNA Nucleoside Deoxyribose Thymine Nucleotide Deoxyribonucleotide Topoisomerases Pentose sugar DNA Transforming Principle Phosphate Group Euchromatin Heterochromatin Phosphodiester Facultative heterochromatin Bond Guanine ADDITIONAL PROBLEM SET: DNA STRUCTURE 1. What are the complementary strands of each of the following DNA strands?. a. 5' AATCGCCCATTGCAGTTC 3' b. 5' CGATTGGCTTA 3' 2. Heat (about 90°C) applied to double-stranded DNA causes the hydrogen bonds holding the strands together to break down. This process resulting in the separation of the strands is known as denaturation. The greater the number of hydrogen bonds in a duplex, the greater the amount of thermal energy required for the denaturation. Therefore, strands with high content C G bases, bases which are held together by three H bonds, will require higher heat to denature than those strands having high content of A T bases , bases which are held together by only 2 H bonds.. The temperature at which half of the hydrogen bonds in a sample of duplex DNA have denatured is called the melting point, or tm~ Listed below are the base compositions of three different DNA molecules. (1) 5' ATTCTAACTTTGAT 3' 3' TAAGAT TGAAACTA 5' (2) 5' CGCACTGGCTAACC 3' 3' GCGTGACCGATTGG 5' (3) 5' CGCAT TAT TGTCAA 3' 3' GCGTAATAACAGTT 5' a. Order these three molecules in terms of their melting points from highest to lowest. 3. Analysis of base composition of DNA molecules from two bacterial strains indicates that the four bases A, T G and C occur with identical frequencies in the DNA from each strain. Does these bacteria strains have identical DNA molecules? 4. Analysis of a nucleic acid molecule indicates that the base thymine is present. No other information is available about the structure or composition of the molecule. a. What kind of sugar would you expect to find in the nucleotides of this molecule? b. What other kinds of bases might be found in this molecule? c. If it is established that thymine makes up 13% of the bases in the molecule and that the molecule is double-stranded, what are the percentages of the other bases? d. If it is established that thymine makes up 13% of the bases in the molecule and that the molecule is single-stranded, can you predict the percentages in which the other bases would occur? Explain. 5. Two DNA molecules, each consisting of 25 base pairs, have the following sequences. 5’ ATATCGGGCTATGAGTACAGTAATG 3' 3' TATAGCCCGATACTCATGTCATTAC 5' 5' AGCATTTCTGACTAATGTACACACC 3' 3' TCGTAAAGACTGAT TACATGTGTGG 5' a. Calculate the A/T ratio for each molecule. b. Calculate the G/C ratio for each molecule. c. Calculate the (A + T)/(G + C) ratio for each molecule. d. Which of the three ratios will allow you to known whether the two molecules have the same overall nucleotide composition ,in spite of having different sequences?. 6. Nucleic acid molecules have been isolated from seven types of viruses. The base composition of these molecules is shown in the table below but the data are incomplete regarding the composition of the nucleic acid, which is given in terms of the percentage of total base composition. Virus Type Base 1 2 3 4 5 6 7 A T C G U 12 12 38 38 0 – 18 – – – 20 0 30 30 20 18 26 26 30 0 – – – – 17 26 0 24 26 24 23 18 28 – – Identify the type of nucleic acid found in each of the seven viruses, indicating, if possible, whether each molecule is single-stranded or double-stranded. Explain the reasons for your classification. 7. Refer to the information in problem 6. Further study of the nucleic acid found in virus 1 indicates that the molecule contains 4200 base pairs. a. How many nucleotides are present in the molecule? b. How many sugar molecules are present in the nucleic acid molecule? c. How many complete turns would occur in the configuration that this molecule assumes? d. What is the length in nanometers of this molecule? 8. Refer to the information in problem 6. Further study of the nucleic acid molecules present in viruses 3 and 4 indicates that the total number of nucleotides present in each molecule is the same. Would these molecules have the same or different lengths? Explain. 9. 1 gram of nucleic acid contains 2.0 x 1021 nucleotides and 2.9 x 106 nucleotide pairs equals 1 millimeter of duplex DNA. The weight of DNA carried in a single genome is called the C-value. In humans this amounts to approximately 3.2 x 10-12 grams. b. Identify the number of nucleotide pairs present in this DNA. c. What is the total length of the DNA in this human haploid cell? Answers: 1a. 3' TTAGCGGGTAACGTCAAG 5' 1b. 3' GCTAACCGAAT 5' 2a. Each duplex has the same number of base pairs. Since molecule 2 has nine CG pairs, it will have the highest tm; molecule 3, with five pairs, will have an intermediate tm; and molecule 1, with three pairs, will have the lowest tm. 3. Not necessarily, the bases may be arranged in different sequences. 4. a. The presence of thymine indicates that this is DNA, therefore the nucleotides would contain the sugar deoxyribose. b. Adenine for sure, although it is likely that guanine, and cytosine would be also present. d. No. The bases in one section of a single strand have no relationship to other bases in the same strand. 5. In both molecules: a. A/T is 15/15 = 1. b. G/C is 10/10 = 1. c. (A + T)/(G + C) ratio for each molecule (15 + 15)/(10 + 10) = 30/20 = 1.5. d. The identical (A + T)-to-(G + C) ratios for each molecule tell us that they have the same nucleotide composition. 6. Virus 1: Since there is no U present, it may be assumed that the molecule is DNA. A = T and C = G, so it must be double-stranded. Virus 2: Since T is present, the molecule is DNA. Lacking information about the frequencies of the other bases, we cannot tell the number of strands in the molecule. Virus 3: Since U is present it must be RNA. A = U and C = G so it must be doublestranded. Virus 4: Since T is present, the molecule is DNA. A does not equal T and C does not equal G, therefore the molecule is single-stranded. Virus 5: Since U is present, the molecule is RNA. Lacking information about the frequencies of the other bases, we cannot tell the number of strands in the molecule. Virus 6: Since U is present, the molecule is RNA. A does not equal U and C does not equal G, therefore the molecule is single-stranded. Virus 7: Since T is present, the molecule is DNA. A does not equal T and C does not equal G, therefore the molecule is single-stranded. The percentage of G can be determined by subtracting the sum of the percentages of the other three bases (23 + 18 + 28 = 69) from 100%: 100% - 69% = 31%. 7. a. 4200 x 2 = 8400 bases for a total of 8400 nucleotides. b. Each of the 8400 nucleotides has one sugar, therefore there are 8400 sugars. c. There is a complete turn of the double helix every 10 base pairs, therefore there would be 4200/10 = 420 turns. d. Each turn has 3.4 nanometers. With 420 turns, the molecule would be 420 x 3.4 = 1428 nanometers long. 8. They have different lengths because virus 3 is double-stranded and virus 4 is singlestranded. Since each has the same number of nucleotides the length of each strand of the double stranded molecule in virus 3 is half the length of the strand in virus 4. 9. a. Number of nucleotides:: (3.2 x 10-12 g) (2.0 x 1021) = 6.4 x 109 nucleotides. b. (6.4 x 109)/2 = 3.2 x 109 nucleotide pairs. c. (3.2 x 109)/(2.9 x 106) = 1.103 x 103 millimeters = 1.103 meters.