Introduction
advertisement

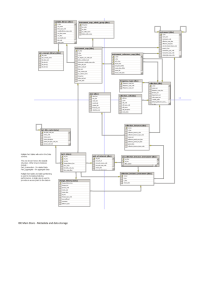
Generic format of the Feasibility Study chapters Introduction While a vast amount of information about the environment and biodiversity can be found in various locations around the world, the comprehensive gathering, organisation, evaluation and dissemination of such information is a challenge which can be addressed only by a variety of distributed and centralized, interoperable and connected services. Such systems should be web based and provide a common framework for the management of this information. In this chapter, we describe the requirements for the effective and efficient integrated management of the existing biodiversity resources and the creation of a common e-services based infrastructure supporting the access, organisation, evaluation and disseminating of such data. The existing biodiversity resources include the entire spectrum from research notes to paper publications, from local IT systems and disparate files to databases accessible on the Web, from observatories to specimen collections and microbiological samples, from species catalogues to –omics databases, and from experimental software packages to Web-services with standard interfaces and behavior. We classify the users into three major groups. These are: End Users (e.g., general public, policy maker, industry) need to locate and extract data that matches their interest, or appropriate data servers to retrieve data of the desired level of quality. They put a high value on the accessibility, interpretability, and usefulness of data. Brokers (e.g., environmental scientists, public authority administrator) maintain secondary information services for end users or answer administrative inquiries, maintain or write new software to access measurement databases, remote sensing data, to build geographical databases to construct maps of a subject and to improve the reliability of data using consolidation techniques. In the most cases these programs may use multiple data sources. Each data source requires a unique program to extract the data for the new program from the data source. Data and service Providers (e.g., biologist, geologist, physicist, ornithologist, entomologist, etc.) collect empirical data, evaluate them and provide access to them for interested parties or the public. This task includes standard database functions or database services fed and updated by automatic sensors that directly transmit their data into them as well as quality assurance of data. They maintain or write new software ranging from standard data evaluation to innovative research tasks. Although Helbionet infrastructure is focuses primarily on the needs of biologists and biodiversity professionals of the research community, it will be geared to supporting a much wider range of users which produce biodiversity information or for which biodiversity information is relevant.. Such uses include: Industry involved in agriculture, fishing, industry under environmental control, political decision support, environmental management and control, support of other European research infrastructures in the biodiversity domain. This chapter will examine the technological aspects of a biodiversity infrastructure for accomplishing the following functionality: - Finding aids: Capability to obtain comprehensive information about all information sources and services, their locations and access methods that may be relevant for a particular user question or research problem via one virtual homogeneous access point based on centralized metadata repositories. This includes electronic records and Web services, but also paper archives and publications, and databases opaque to the Web. - Centralized Service-directory services: services describing content of and access to electronic collections and other Web-services in a way that compatible ones can be identified and be connected in scientific workflows. - Access: Ability and permission management to access or download all relevant information that 1 exists in electronic form from any location with Internet access. Ability to mediate or transform between major standard data formats. Ability and permission management to invoke Web-services for complex evaluation tasks, including ability of interlinking automated observatories Semantic interoperability and description: Homogeneous core metadata capturing the key concepts and semantic relationships to characterize the relevance of a data set or service for an information request or research problem, such as correlating professional scientific or amateur observations with species, habitat and molecular data and various theories on the behaviour and evolution of biodiversity, in particular in order to give answers for complex decision problems in environmental policy. Managing the dynamic resolution and curation of co-reference, i.e. linking identical resources and references to identical resources referred to by different names or identifiers, thus leading to an all-increasing state of semantic connectivity of the distributed resources in the infrastructure. - - Scope The semantics of the biodiversity universe of discourse can be characterized by the following core conceptual model of fundamental categories and their relationships. It is a high level picture of the relationships between human activities and their targets, as they appear on the highest level of description of all scientific products and services. They are the ones necessary for the first level selecting relevant information, and for managing and maintaining the referential integrity of the most fundamental contextual information. It is only on a more specialized level that all the entities in these models are refined by specific, open-ended terminologies (typologies, taxonomies) and a relatively small set of more specific relationships among them: services Molecular world and parts global indices databases Publications exemplifies Samples use to appear in is about from exemplifies Species specimen Individuals/ treat use cross reference in collections in from appear in create maintain Records Activities Habitats Observations Place Simulation Time Evaluation The general idea is that biodiversity Activities (like Observations, Simulations, Evaluations etc) of human Actors deal primarily with individual items or specimen coming from habitats or laboratory and occur in a particular space-time. Evidence of these activities is produced in the form of physical collections of items (green) and information objects (blue) which are about themselves and the observed world. The biodiversity world produces categorical theories (yellow) on the species and population level and on the 2 associated micro- and molecular phenomena, and regards primary observations and physical items and examples of those theories. The data records are stored in information systems accessible via services. Global indices can be created for accessing them comprehensively. The data bases also keep information about species, activities, habitats, etc. (Generically, we regard a habitat as a set of coherent physical phenomena and their constituents that can be observed over a specific place and time period). Biodiversity research functions may include: species declaration, determination and other kinds of scientific classification, , biodiversity monitoring, hot-spot comparison, , phylogenetic and bio geographic analytical tools, virtual population analysis tools, simulation and prediction tools, multivariate analyses, geospatial management etc. In the above world of biodiversity we consider different levels and kinds of metadata according to the nature of the described units and the way they can be accessed. These are: - Data record metadata Collection-level metadata describing a digital collection or part of it as one unit. Metadata of live digital sources (services providing records from automatic sensors or regularly updated databases ) Metadata of computational services and tools. Collection-level metadata describing a physical collection or part of it as one unit. Physical collection item metadata (specimen, samples) Paper publication metadata (library records) Metadata about paper archives describing a paper/photo collection or part of it as one unit. The above conceptual model will be refined and used for providing access points and cross-linking for all the above metadata. In addition, central semantic knowledge services are needed to maintain a global notion of identity about key national people (see www.viaf.org), activities, habitats, places (georeferencing services), with suitable linking into respective international resources. Following the conceptual model above, these services are needed to connect the source metadata with clear-cut identification of the associated contextual elements of observations, on which their understanding and interpretation ultimately relies. In this generic conceptual framework, a state-of-the-art management of distributed, interlinked terminologies (“ontologies”) plugs in. State of the Art, Currently there is a lot of effort on distributed systems that handle massive data load in conjunction with semantic information on distributed content. On one side, there is the huge state-of-the-art of the bioinformatics world, including global integration efforts of molecular biology data (“omics”), specimen holdings and species catalogues. It is not the aim of this infrastructure to replace or improve these efforts, but to connect such kinds of services and to complement these services with an even more generic global directory service, which, in the sequence, allows to maintain “Data Cloud” and “data GRID” like structures. State-of-the-art biodiversity services appear in this architecture as components maintained by disciplinary subgroups of the infrastructure users. It is just very recently (about 2 years) that scalable RDF-triple store solutions allow for implementing very large integrated metadata repositories with deep referential identity and integrity, and therefore tracing related resources across all kinds of sources and services and by rich contextual associations that could never before established in this comprehensive form. It should become clear, that an integrated semantic access as outlined above is fundamentally different from Web search engines. The success of the latter is based on the fact that information about some key concepts is highly redundant on the Web. Therefore, users can easily be guided towards some relevant content. In a descriptive science as biodiversity, users look for facts that have no name as such and are observed only once. To enable such research, we need semantic networks of deeply linked information to enable targeted navigation through related contexts and factors. Not a “see also”, but a “found individuals of species X in place Y in individuals of species Z”. 3 Here some of the most innovative examples of such semantic integration systems are presented. The Information Systems Laboratory (ISL) of FORTH and its Center for Cultural Informatics have participated in one way or another in many of these, and accumulated a considerable know-how. Some of these are directly in the biodiversity domain, but others are more generic, or in the cultural heritage domain. Cultural heritage and biodiversity have a lot in common, as both deal with records of recent and past observations and their interpretation, and holdings of physical collections. Indeed, FORTH has leaded a working group that identified common core models for biodiversity museums and cultural museums. We present here some recent national and international projects that relate to the state-of-the-art of the technology we suggest to provide central services to an integrated, distributed biodiversity infrastructure in Greece and its challenges: LIFEWATCH LifeWatch will be a distributed research infrastructure, where the entities of the LifeWatch organization and the services that the LifeWatch organization provides will be spread over Europe. This research infrastructure is the permanent elements needed to create an internet/web-based inter-organisational system that links personal and institutional systems (data resources and analytical tools) for biodiversity research, supported by appro-priate human resources providing assistance to users. The proposed architecture of the HELBIONET infrastructure is compatible with the proposed LIFEWATCH Architecture. The main elements of the proposed Lifewatch architecture are: - The Resource Layer contains the specific resources, such as data repositories, computational capacity, sensor networks / devices and modelling / analysis tools, that contribute to the Life-Watch infrastructure. The primary components at this layer are the biological (e.g. species re-cords) and abiotic data from sites. Additional components include catalogue services (e.g., taxo-nomic checklists or gazetteers), analysis tools, and processing resources. - The e-Infrastructure layer provides mechanisms for sharing the specific resources as generic services in a distributed environment, spread across multiple administrative domains. This in-cludes systems for identifying, accessing, and processing resources located within multiple ad-ministrative domains, uniform security and access protocols, and ontologies for semantic integration. - The Composition Layer supports the intelligent selection and combination of services in order to complete tasks. This includes workflows, semantic metadata for the discovery of components and the storage of additional attributes such as provenance and version information. Viewed from the perspective of the biodiversity scientist, the Composition Layer consists of a wide range of application services or biodiversity research capabilities that can be selected and arranged in an infinite number of ways to conclude specific analytical and experimental tasks. - The User Layer provides domain-specific presentation environments for the control and monitor-ing of tasks and tools to support community collaborations. This includes a LifeWatch portal incorporating discovery and workflow management tools and offering a single point of access for all users. Institute of Museums and Library Services, Digital Collections and Contents (IMLS DCC) <http://imlsdcc.grainger.uiuc.edu/about.asp> The Digital Collections and Content (DCC) developed database about digital collections (some 900). Each collection is described by collection level metadata. In this project a logic-based framework for classifying collection-level/item-level metadata relationships is developing. The idea behind this framework is very similar to what we propose for HELBIONET for the Data Dictionary services. This framework supports (i) metadata specification developers defining metadata elements, (ii) metadata librarians describing objects, and (iii) system designers implementing systems that help users take advantage of collection-level metadata. The users can be search for collections and in collections. Metadata relationships for collection/item and metadata at the collection-level are classified and are used for finding, understanding, and using the items in the collection. Much effort has been made to map between different metadata schemas used by many different communities. In the next phase of development the employment of CIDOC – CRM is considered. CLassical Art Research Online Services (CLAROS), http://www.clarosnet.org/about/default.htm CLAROS is an international interdisciplinary research initiative. There are five founder members of CLAROS, each with extensive datasets on classical antiquity (mainly art) that have been created over the past thirty years. 4 Collectively they have more than two million records and images that will be available through CLAROS. The goals of CLAROS are very similar to the goals of HELBIONET about the persistent data. The user communities are different since CLAROS addresses the needs of the art world. The architecture of ClarosWeb is very similar to what we propose for HELBIONET. It is employing mapping, coreference, query, search and browsing, and multilingual thesaurus services. Additional classical art data providers can be integrated into the system at any time, thereby enriching the total content, simply by mapping their schemas to the core ontology and by making appropriate entries in the co reference service. Each member retains his own assets in his own format, his own website and his own IPR . CIDOC-CRM has been adopted for core ontology since CIDOC-CRM allows common data elements to be used to represent common concepts, without constraining the overall range of information that can be represented. This ontology form the foundation for a service that indexes key concepts from these sources to provide a query facility that operates uniformly across all these data. CIDOC-CRM also supports the targeted use of thesauri for multi-language support, both in the diversity of data sources and in user queries. CultureSampo –A National Publication System of Cultural Heritage on the SemanticWeb 2.0 http://www.kulttuurisampo.fi/index.shtml CULTURESAMPO is an application demonstration of a national level publication system of cultural heritage contents on the Web, based on ideas and technologies of the Semantic (Web and) Web 2.0. The material consists of heterogeneous cultural content, which comes from the collections of 22 Finnish museums, libraries, archives, and other memory organizations, and is annotated using various ontologies. All of these annotations are represented using the Resource Description Framework (RDF) and a set of ontologies, including the Finnish Spatio-temporal Place Ontology SAPO. The material, coming from almost 100 different collections, currently includes metadata about over 250,000 objects, e.g. artefacts, photographs, maps, paintings, poems, books, folk songs, videos, et cetera, and millions of other reference resources (concepts, places, times, etc.). CULTURESAMPO provides a centralized geospatial view that can show all kinds of content related to a place, and organize this data in various ways in order to make sense of it. It employs Google maps view, upon which are shown by default all locations to which any content is related in any way. It provides a search box, whereby the user can search for places by name using semantic autocompletion, and a browsable partonomy of places in the ontology. The common approach of this project with the HELBIONET node is the usage of semantic models based on events for addressing the interoperability problems and geospatial models for addressing a geospatial view. Linking Open Data (LOD), http://esw.w3.org/SweoIG/TaskForces/CommunityProjects/LinkingOpenData The Linking Open Data (LoD) project, an open, collaborative effort carried out in the realm of the W3C SWEO8 Community Projects initiative aimed at bootstrapping the Web of Data by publishing datasets in RDF on the Web and creating large numbers of links between these datasets. Currently, the project includes over 50 different datasets with over two billion RDF triples and three million (semantic) links at the time of writing | representing a steadily growing, open implementation of the linked data principles. RDF links enable the navigation from a data item within one data source to related data items within other sources using a Semantic Web browser. RDF links can also be followed by the crawlers of Semantic Web search engines, which may provide sophisticated search and query capabilities over crawled data. As query results are structured data and not just links to HTML pages, they can be used within other applications. Collectively, the data sets consist of over 13.1 billion RDF triples (dataset statistics), which are interlinked by around 142 million RDF links (linksset statistics) (November 2009). In HELBIONET infrastructure the terminologies and georeferencing will all be connected via LoD. 3D-COFORM, www.3d-coform.eu The 3D-COFORM integrated infrastructure allow different classes of users (CH professionals, technicians, academics, museum users and web users, i.e. European citizens) to create 3D collection items by digitizing, processing, classifying, annotating, cross-referencing and storing digital objects with rich semantics into a repository. It employs ontologies to ingest, store, manipulate and export complex digital objects, their components and related metadata as well as to enable efficient access, use, reuse and preservation. The CIDOC CRM is used as a common starting point in 3D-COFORM for data interoperability between the partners and supporting Cultural Heritage institutions in the consortium. Each gallery or museum schema is be mapped to the CIDOC CRM and the development of extensions in collaboration with the standards bodies where necessary (e.g. the current work of the community on an extension for recording digital provenance). The decision to target the 3D-COFORM interoperability over a CIDOC CRM structured collection is a significant and important point, and carries with it the requirement to ensure compatibility between a CIDOC CRM orientated 3D-COFORM repository and other interoperable digital libraries like Europeana, which may have 5 initially targeted other approaches. The CCI of ISL of FORTH design of the 3d-coform repository infrastructure (RI) and design and implement the metadata repository. Metadata are ingested in the repository as RDF files. The components supply the RI with RDF or XML files compliant to a specific Schema, which is produced based on the extended CIDOC-CRM model. XML files will be internally transformed to RDF files in the RI. Also CCI developed the appropriate provenance model for digital objects that is more rich than the existing proposals. It employs the notion of a digital machine event, digital measurement and formal derivation are very generic, and the essence of e-science. The notion of digitization is specific to certain processes, and assists reasoning about “depicted objects”. Similar specializations may be created to reason about other measurement devices. In the Helbionet we propose the same technology for the repository infrastructure and the employment of the same provenance model. CIDOC CRM, http://cidoc.ics.forth.gr/ The CIDOC Conceptual Reference Model (CRM) provides definitions and a formal structure for describing the implicit and explicit concepts and relationships used in cultural heritage documentation. The CIDOC CRM is intended to promote a shared understanding of cultural heritage information by providing a common and extensible semantic framework that any cultural heritage information can be mapped to. It is intended to be a common language for domain experts and implementers to formulate requirements for information systems and to serve as a guide for good practice of conceptual modelling. In this way, it can provide the "semantic glue" needed to mediate between different sources of cultural heritage information, such as that published by museums, libraries and archives. The CIDOC CRM is the culmination of over 10 years work by the CIDOC Documentation Standards Working Group and CIDOC CRM SIG which are working groups of CIDOC. Since 9/12/2006 it is official standard ISO 21127:2006. Besides others, the Centre works as competence center for the CIDOC-CRM (ISO 21127), by building up and exchanging application know-how, consultancy to implementers and researchers, and contribution to the dissemination, maintenance and evolution of the standard itself. EUROPEANA, www.europeana.eu Europeana rolls multimedia library, museum and archive into one digital website combined with Web 2.0 features. It offers direct access to digitised books, audio and film material, photos, paintings, maps, manuscripts, newspapers and archival documents that are Europe’s cultural heritage. Visitors to www.europeana.eu can search and explore different collections in Europe’s cultural institutions in their own language in virtual form, without having to visit multiple sites or countries. The digital objects that users can find in Europeana are not stored on a central computer, but remain with the cultural institution and hosted on their network. Europeana collects contextual information about the items, including a small picture. Users will search this contextual information. Once they find what they are looking for, a simple click provides them with access to the full content – inviting them to read a book, play a video or listen to an audio recording – that is stored on the servers of the respective content contributing institutions. The Europeana prototype gives direct access to more than 2 million digitised items from museums, libraries, audiovisual and other archives across Europe. Over 1,000 cultural organisations from across Europe have provided materials to Europeana. The digitised objects come from all 27 Member States, although for some of them the content may be very limited at this stage. The HELBIONET infrastructure has similar functionality with Europeana. Martin Doerr, head of the Center of Cultural Informatics of ICS of FORTH is core a expert of Europeanna and has contributed a larger part of the EDM model, the schema of the next (“Danube”) release of Europeana. This model is a generalization about CIDOC CRM, DC, and ORE. ACGΤ (Advancing Clinico-Genomic Clinical Trials on Cancer - IST-026996) ACGT brings aims to deliver to the cancer research community an integrated Clinico-Genomic ICT environment enabled by a powerful GRID infrastructure. In achieving this objective ACGT has formulated a coherent, integrated work plan for the design, development, integration and validation of all technologically challenging areas of work. Namely: (a) GRID: delivery of a European Biomedical GRID infrastructure offering seamless mediation services for sharing data and data-processing methods and tools, and advanced security; (b) Integration: semantic, ontology based integration of clinical and genomic/proteomic data - taking into account standard clinical and genomic ontologies and metadata; (c) Knowledge Discovery: Delivery of data-mining GRID services in order to support and improve complex knowledge discovery processes. The technological platform will be validated in concrete setting of advanced clinical trials on Cancer. Pilot trials have been selected based on the presence of clear 6 research objectives, raising the need to integrate data at all levels of the human being. ACGT promotes the principle of open source and open access, thus enabling the gradual creation of a European Biomedical Grid on Cancer. Hence, the project plans to introduce additional clinical trials during its lifecycle The Information Systems Laboratory of ICS develop ed, revised and extended, in collaboration with IFOMIS, the ACGT master ontology (MO). The intention of the ACGT Master Ontology (MO) is to represent the domain of cancer research and management in a computationally tractable manner. Also ISL work on - the design the ACGT mapping format; - the specification of requirements and the evaluation of the following semantic integration tools (developed by UPM): The mapping tool used to map the sources to the MO The ACGT mediator used to integrate clinico-genomic information. - The development of the Ontology Submission tool: a tool that intends to aid the evolution of an ontology through requests for change from users and domain experts. The submission tool was integrated to the Trial Management system (Obtima) using web services. Mapping the master ontology of ACGT(MO) to the CIDOC CRM important relationships about scientific observation extracted from the CRM. As a result we could say that the CRM with minor extensions can be a core model to integrate clinical observation and Omics laboratory measurements. POLEMON The system POLEMON, a decentralized management information system for the National Monuments Record funded by EPET II program by GSRT, has employed a directory service on a distributed network of cultural information databases. This system was designed for the Greek Archeological Service, one of the oldest public services in Greece, was organized from the very beginning decentralized. The peripheral units, called Ephorates of Antiquities, approach now 67 in number and include a number of large independent Museums and some special services (Underwater Archaeology, Speleology etc.,) are responsible for the field work, excavations, restorations, protection and management of the archeological sites and monuments and their environment . A special Direction at the General Directorate of Antiquities and Restoration, called Directorate of Monuments Records and Publications collects any useful documents concerning sites and monuments, compiles a general inventory and classifies the historic archives 160 years old of the Greek Archeological Service. The peripheral units may have their own database management system to manage their monument data and they give by request permission to researchers to access this data. These dispersed isolated cultural databases along the Ephorates may have the same logical schema or not as well as their vendors and type may be varied and may concern different types of monuments. The CCI of ISL of FORTH was the coordinator and responsible for the design and implementation of the global access system (directory service) of POLEMON. The exploitation of the POLEMON system has started since 1997 with the participation of 12 peripheral units of the Ministry of Culture. The current installation of POLEMON system in Ministry of Culture employs 67 dispersed databases over the Greece. The metadata repository of the Global Access System of POLEMON is very similar with the metadata repository which we propose for HELBIONET infrastructure. Also the mapping method of different databases schemas to a global schema is very similar to what we propose for the HELBIONET mapping mechanism. German Federal Ministry of Education and Research (BMBF): Research Between Natural and Cultural History Information: Benefits and IT-Requirements for Transdisciplinarity This research was partly supported by the German Federal Ministry of Education and Research (BMBF) within the BIOLOG Biodiversity Informatics- and BIOLOG Biota Africa- programmes by funding the DORSA-, DIG- and BIOTA East Africa E15 projects. http://doi.acm.org/10.1145/1367080.1367 The core ontology we propose to HELBIONET is very similar with the one employing to this research. The adopted approach in this research is to integrate into core ontology transdisciplinary information. The proposed core ontology is based on the CIDOC-Conceptual Reference Model (ISO 21127). When the proposed ontology is instantiated with some realistic examples taken from the field of biodiversity (collecting, determination, type creation, expedition, and observation events), the formal specification of semantic concepts makes scientific activities commonly understandable. This research proved that ontologies not only allow one to describe the results of scientific activities, such as a description of a biological species, but they can help to clarify the path by which the goal was reached. In particular, ontologies provide a high-level uniform representation of transdisciplinary research activities and results. Also it is proved that ontologies as knowledge representation tools will have strong impact on methodological questions and research strategies for different 7 domains such as biology, archaeology, art history, and socio-economy. They can be regarded as semantic glue between and within scientific and scholarly domains as demonstrated in a series of examples. Requirements In the light of existing infrastructure in Greece, a general requirement is posed for the design and implementation of a directory service for data and service discovering The proposed infrastructure should support biodiversity research functions for the three types of user groups (End Users, brokers, data providers). The list of required functionality can be grouped according to the type of the user group. The end users need functions for discovering, accessing, processing and viewing data. These functions include: Searching and browsing mechanisms for distributed data and services. Uniform identity framework for data and services Access to existing data and services, distributed among multiple organizations Mechanisms for source data preservation, i.e., access to past versions of data sets that have been used to produce secondary information. Mechanisms for data analysis as well as mapping and modelling tools, using standard ways to manipulate and view data. Mechanisms for data fusion, integrating different sources (such as sensor data, biodiversity parameters, geographic data, primary data, workflow execution), to allow fast retrieval at different levels of detail e.g. for analysis and visualization. Support the understanding of results by the user, by providing tools and mechanisms to enhance knowledge extraction from discovery as well as from analysis results The brokers need functions for composing electronic workflows. These functions require translation or mapping mechanisms between data formats and semantics. Broker functions include: Mediation mechanisms like a semantic metadata framework should enhance interoperability between resources. Promote interoperability standards that will allow characteristics user groups to combine particular services such that they can support for collaborative workflow modelling, providing documentation and enactment of data processing steps. management of provenance metadata information and promotion of provenance metadata generation by data providers following adequate standards. Mechanisms for managing data sets and their processing tools independently by their providers and mechanisms for reproducing or examining an analysis of a specific version of both the data and the tools used in this analysis. Support for building and managing virtual organisations by allowing the definition of groups and roles and their authorisation for sharing resources Data and service providers continue to manage their data (and services) independently as now, including control of the creation and modification of data/services. However, data can be accessed by authorised users located anywhere through a generic mechanism defined by Helbionet. They need functions for Capturing data from users and lightweight devices, including field sensors and networks providing continuous streams of new data, and portable computing devices, often with intermittent connectivity. Allowing users to add annotations to existing data and services, which may contribute to the quality assessment and feedback processes; Providing control mechanisms for access to data and services, together with monitoring services for the support of service level agreements and, potentially, including charging mechanisms . From the technical point of view the requirements include: Evolutionary development Clear separation between centralized services and local data and service provider responsibilities. High on-line accessibility of all data Technology independence and rigorous use of standards Metadata information about any source of data independent of their format and age Metadata integration and central directory services (integrated access to collection level and item level metadata) 8 Loosely coupled components Generic, self-describing infrastructure components Distributed systems interoperability Unambiguous identification (identifier generation service) Dynamic co reference (identity) resolution of URI’s, links, identifiers to: species, specimen, samples, data records, observation campaigns, publications, archives of scientific work records, services and software tools, places, actors, habitats All the above requirements are preconditions which should be followed in national level, in the forthcoming phase of the project, in order to be accomplished tighter integration of tools and services in a next step of this project. A tighter integration of tools and services include Support workflow modelling, providing documentation and enactment of data processing steps. Collaborative community support tools which allow groups of people to work on a shared objective. Recommendations and conclusions In the light of this study, we may say that the Greek HELBIONET infrastructure should have the character of a directory service for data discovery and access and interoperability promotion. Also substantial access services will be provided in the form of Linked Open Data (LoD). The architecture of the national node The service-oriented architecture (SOA) is an appropriate high – level architecture for the HELBIONET infrastructure to follow. A deployed SOA-based architecture will provide a loosely-integrated suite of services that can be used within multiple biodiversity domains. SOA defines the interface in terms of protocols and functionality and separates functions into distinct units, or services, which developers make accessible over a network in order to allow users to combine and reuse them in the production of applications. SOA implementations rely on a mesh of software services. Services comprise unassociated, loosely coupled units of functionality that have no calls to each other embedded in them. Each service implements one action. Instead of services embedding calls to each other in their source code they use defined protocols that describe how services pass and parse messages, using description metadata. These metadata should describe in sufficient detail not only the characteristics of these services, but also the data that drives them. Extensive use of XML is made in SOA to structure data that they wrap in a nearly exhaustive description-container. Analogously, the Web Services Description Language (WSDL) typically describes the services themselves, while the SOAP protocol describes the communications protocols. The great promise of SOA suggests that the marginal cost of creating the n-th application is low, as all of the software required already exists to satisfy the requirements of other applications. Ideally, one requires only orchestration to produce a new application The global system of the Greek HELBIONET infrastructure will constitute a directory service of data and services. The data and service providers are responsible to manage their products (data and services) independently (including the creation and modification of data/services) and make public the products they want. Services on providing data can be distinguished in (a) persistent data services, the services that provide access to database data, (b) live services, that provide dynamic data by accessing data generating devices, (sensors, metering stations, etc.), and (c) computational data services, that given a set of data inputs they produce a set data outputs (simulation data). Only the persistent data services provide data in the form of Linking Open Data (LoD), and thus metadata on these data can be harvested. This directory service will provide to the data/service providers the way to declare themselves as providers, to declare the services and data they provide and to publish (or update the already published) data and services. All providers should declare their services following the directory provided WSDL description. The same holds for all the data which the providers wish to make public. First they have to declare the kind of data they wish to provide, so the directory is aware of what kind of data is available from each provider, and thus redirect the users to the specific provider. The providers should follow a specific directory provided WSDL description for 9 publishing their data. They have to provide their data as a data-service, implementing a Web-Service on top of their databases, and to declare this data service to the directory. For being accessible the data of different providers through a central search mechanism, the providers should also supply the metadata of this data. All appropriate semantic information (metadata) should be defined in advance in order to facilitate the querying and retrieval process. Data providers should also provide a metadata extraction service that will allow the directory service to harvest the available metadata of the data of a provider’s database. All harvested metadata will be mapped from their original XML format to a unified and normalized XML schema which will be based on CIDOC – CRM (CRM core schema compatible). This unified schema will contain all the well-defined mappings of the different databases schema to this normalized schema. All metadata will be stored in the metadata repository. The metadata XML original format should be compatible to a specific set of XML schemas and DTDs (according to the international standards which are being used in these fields, e.g. ISO 21127, DUBLIN CORE, EAD). In parallel the system will trace and retrieve terms from metadata that will be co-referenced with external authorities. For example geographical locations will be co-references with geographical dictionaries or directories (gazetteers). Thus by ingesting the metadata the system will also ingest terminology that will be used by the search mechanism of the system. The system should be globally accessible via a web interface (and a set of Web-service actions) which will enable query formulation on data, metadata, services, in conjunction with the terminology manager in order to provide successful query results. In light of the above, the description of the structural and functional components of the proposing system follows: Functional components The system consists of the following functional components: Directory of Providers Services Responsible for creation, deletion, manipulation, management of services from all providers. Constitute the central directory the external users will access in order to get information on available data, or available data and/or services Provide the directory of all the providers Metadata repository Responsible for creation, deletion, manipulation, management of the metadata that the providers would like to be public. Unambiguous identification (identifier generation service) Dynamic co reference (identity) resolution of URI’s, links, identifiers to: species, specimen, samples, data records, observation campaigns, publications, archives of scientific work records, services and software tools, places, actors, habitats 10 HELBIONET Directory service system Access Web-service interface Query Result Management Access Management Query interface output interface Metadata Repository Application server Providers and Services retrieval interface Applicati co-reference on server directory Directory of Providers and Services Input interface output interface Metadata loading interface output interface Application Terminology server extractor Providers and Services input interface Input interface Application Mapping server mechanism Input interface Ingest Web-service interface Ingest tool sources Declaration of Services and Data Meta data Web-Service interface Local Data Base Declaration of Services and Data Meta data Declaration of Services and Data Meta data Web-Service interface Web-Service interface Local Data Base Local Data Base Declaration of Services and Data Meta data Web-Service interface Local Data Base Mapping mechanism Provides the mechanism for mapping the metadata from the data files described in various formats (ISO 21127, DUBLIN CORE, EAD) to the unique format accepted by the Metadata repository (CIDOC - CRM core schema compatible) Co-reference directory server Adds/deletes/changes terms and their co-references in externals authorities. Maintain relations between terms Create/delete/maintain term hierarchies Queries on term co-references and their relations Terminology extractor Extracts terms from text (metadata or documents) Search in external authorities for co-referencing information on the extracted terms. Co-reference directory controller Implements all the logic and control on extracting terminology. Stores and queries the extracting terminology. Works like a “glue” between the co-reference dictionary and the terminology extractor. Provides the interface for both (i) input mechanism of the co-reference directory server: [gets a text (in a buffer or in a document) calls the terminology extractor and extracts the terms and their co-references with external authorities, and stores them in the co-reference directory server], and (ii) output mechanism of the co-reference directory server: [based on a term, returns its co-references with external authorities). Ingest-tool controller Implements all the logic and control for loading the metadata from various selected databases Works like a “glue” between the directory repository, metadata repository and the co-reference directory 11 server, concerning the metadata loading and maintenance When invoked to load a new set of metadata files the following actions will take place: (i) invocation of the mapping mechanism to produce uniform metadata for all input metadata, (ii) creation of the appropriate linking from the metadata to the actual data objects (LoD), (iii) invocation of the coreference directory controller to extract terms from metadata and from any data objects (basically the documents), and store them along their co-references to the appropriate authority entries, (iv) storing the metadata into the metadata-repository. Access manager To get a result from a database, the user may either use predefined queries; either create its own query by querying one or more values on one or more tags in the metadata schema To get the list of services from a provider, to get a service description (what the service is about), to get the method to invoke the actions of a service.. Results presentation mechanism The system will provide mechanisms for presentation and management of the results. These are merging, ranking, exporting and saving results mechanisms. Access mechanism controller Implements all the logic and control for querying and accessing the data from the system. Works like a “glue” between the directory repository, metadata repository and the co-reference directory server, concerning the data and service retrieval and presentation Basically it is controlled by the user who invokes the UI functions, or the web-service caller, in order to make queries to various databases and collect the results. Is invoked to get the list of services from a provider, or the list of providers that may be candidates for the kind of data the user want to query and retrieve, to get a service description (what the service is about), or to get the way the service actions may be invoked. When invoked to execute a query the following steps will take place: (i) a set of queries will be formulated by the unfolding of the terms that are queried in order to get all their co-references, (ii) in some cases the terms and their co-references may be used to get data records from the providers. The system will provide also a co-reference browser in order for the user to get more terms to choose from. The system will provide the following results presentation and management: merging, ranking, exporting and saving results as well as querying on a result set. Investment costs We estimate the following cost for the development phase and for the testing and support phase. Development phase: Workload: The following table presents workload costs: Personnel type Developers Senior researchers senior Engineers Overhead Total number of persons number of years number of months per year total 3 4 12 144 2500 360.000 2 4 6 48 6000 288.000 288.000 1 4 12 48 4000 192.000 840.000 192.000 922.800 138.420 1.061.220 15% monthly cost subtotal FPA (23%) 82800 Subtotal 442.800 S/W: The s/w platform for the development will be based on Open source technology development tools. We anticipate that some of the development s/w may not be OpenSource, but we estimate the cost of such s/w needed not to exceed the amount of ~10.000 euro (s/w for geographical representations, maps e.g. like the development suite of Google-maps, 3D presentations and previewing tools, statistics tools, etc) H/W: For the development of the project we need the 2 development PCs (~1.500 euro) and one server (~ 3.000 euro)(running Windows server or linuX). 12