CPD Course: REAL
advertisement

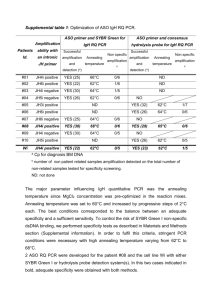
Molecular Biology: Real-time PCR Molecular Biology: Real-time PCR Author: Dr Kgomotso Sibeko-Matjila Licensed under a Creative Commons Attribution license. Pre-requisites for the sub-module on Real-time PCR: 1. General theory on Molecular Biology 2. Polymerase chain reaction (PCR) ANALYSIS Real-time PCR amplifies and simultaneously quantifies a targeted DNA or RNA molecule making it suitable for both qualitative and quantification analysis. Qualitative detection: FAQ11Qualitative detection determines the presence or the absence of target nucleic acid in a biological material. During amplification, the real-time PCR system monitors the accumulation of the PCR product using fluorescence. The fluorescent signal increases proportionately to the accumulated PCR product. When the fluorescent signal reaches detectable levels it is captured by the system and displayed as an amplification curve. Theoretically, the amplicon concentration is expected to increase exponentially during the initial phase of the amplification process (Swillens et al., 2008). At the final phase, the amplification curve deviates and bends toward a plateau, as a result of decreasing DNA polymerase activity or depletion of essential reaction components e.g. primers or fluorescent probe. Consequently, the amplification curve is generated as a sigmoidal shape plotted from fluorescence data vs cycle (Fig. 2). 1|Page Molecular Biology: Real-time PCR Fig. 2: Typical amplification plot. The amplification plot or curve is generated as a sigmoidal shape plotted from fluorescence data against the cycle number. An amplification curve is only produced when target nucleic acid has been detected in a sample (blue curve); in the absence of fluorescence, as is the case where amplification did not occur, a straight line is generated (green line). An amplification curve is only produced when target nucleic acid has been detected in a sample; in the absence of fluorescence, as is the case where amplification did not occur, a straight line is generated. An amplification curve may be observed when there has been contaminating material amplified, resulting in a false positive result. However, FAQ12when using SYBR green and hybridization probes, a melt curve analysis can be performed on the amplification product to confirm if the product is the desired target product (Fig. 3). 2|Page Molecular Biology: Real-time PCR Fig. 3: The melting curve is produced when the fluorescence is plotted against temperature and then the change in fluorescence/change in temperature (-ΔF/ΔT) is plotted against temperature to obtain a clear view of the melting dynamics as shown by the melting peaks on the figure. Based on the Tm of each amplicon produced (as determined by the amplicon sequence composition), the melting curve analysis allows differentiation between specific and non-specific PCR products. FAQ13Melt curve analysis measures the dissociation characteristics of double-stranded DNA during heating. In a SYBR green reaction, the fluorescent signal decreases as a result of the separation of DNA double strands, ultimately releasing the SYBR green molecules. In a hybridization probes reaction, raising the temperature causes the probes to melt off the target product resulting in the separation of the donor and acceptor dye molecules; consequently FRET is reduced and the fluorescence is decreased (www.roche-applied-science.com/lightcycler/). The temperature at which half the FRET signal is lost is referred to as the melting temperature (Tm) of the probe. This temperature varies depending on the DNA sequence, length and GC content. The Tm changes with even a single nucleotide difference. In addition to differentiating between specific and non-specific products, this characteristic of the melt curve analysis allows detection of single-nucleotide polymorphisms (SNP), distinction of homozygous and heterozygous gene alleles by the dissociation patterns produced and discrimination between species of the same genus. During melt curve analysis, the fluorescence is measured continuously as the temperature is increased. The real-time PCR system generates a ‘melting-curve’ by plotting the decreasing fluorescence data against temperature. The real-time PCR detection systems calculate the first derivatives of the curves, resulting 3|Page in curves with peaks at the respective T ms Molecular Biology: Real-time PCR (http://www.qiagen.com/resources/info/guidelines_rtpcr/dataanalysis_sybr.aspx) (Fig. 3). Therefore, PCR products of different sequences have different T ms and products of identical sequences are expected to have the same Tm. A single nucleotide polymorphism in the target DNA under a hybridization FRET probe will still generate a signal and the melting curve will display a lower Tm. Therefore, a different Tm may be an indication of sequence differences under the probes region. When more than three base pair differences occur under a FRET hybridization probe region, hybridization at typical annealing temperatures may be prevented and the products are not detected. Primer dimers generate curves with peaks at a Tm lower than that of the specific PCR product, while non-specific products and smears produce diverse peaks with different Tms. Quantitative analysis: Real-time PCR systems use a fluorescent signal measured during the exponential phase of the amplification process for quantitative data analysis. The amplification curve generated contains valuable information that allows the user to determine the concentration, or relative concentration of target DNA or RNA in unknown samples. To analyze quantitative data, the instrument uses two important parameters: 1. FAQ14The Cycle threshold of the sample Cycle threshold (Ct) is defined as ‘the cycle at which the fluorescence of a sample rises above the background fluorescence’ (www.roche-applied-science.com/lightcycler); it is the intersection between an amplification curve and a threshold line (Fig. 4). At this point, a detectable amount of amplicon has been generated during the early exponential phase of the reaction. 2. The Threshold line The threshold line is the level of detection at which a reaction reaches a fluorescent signal above background. The threshold line is set in the exponential phase of the amplification to allow accurate analysis (Fig. 4) (www.appliedbiosystems.com). Its intersection with the amplification curve determines the Ct, thus Ct = the number of cycles required to reach the threshold. 4|Page Molecular Biology: Real-time PCR Fig. 4: The graphic representation of PCR data showing several parameters of the real-time reaction amplification plot (including the threshold cycle, threshold line and baseline). The threshold cycle (Ct) is the intersection between an amplification curve and a threshold line. It is a relative measure of the concentration of target in the PCR reaction. The threshold must be set in the linear phase of the amplification plot. The Ct value increases with a decreasing amount of template. FAQ15C t values in real-time PCR correlate closely with the original quantity of target sequences and are influenced by the concentration of the target. The Ct value increases with a decreasing amount of target and vice versa. For absolute quantification, external standards of known concentration are used to generate a standard curve from which the concentration of an unknown target can be extrapolated (http://www.qiagen.com/resources/info/guidelines_rtpcr/dataanalysis_sybr.aspx) (Fig. 5). The Cts of the standards are plotted against the log of the template amount, resulting in a straight line. The Ct values and the standard curve are then used to calculate the amount of starting template in an unknown sample (Vaerman et al., 2004; Rutledge and Côté, 2003; He et al., 2002). 5|Page Molecular Biology: Real-time PCR Fig. 5: The amplification plot and standard curve for absolute quantification. External standards of known concentration are used to generate a standard curve from which the concentration of an unknown target can be extrapolated. The Cts of the standards are plotted against the log of the template amount, resulting in a straight line. 6|Page