SEARCHING A FUNCTION FOR PPI1, AN INTERACTOR OF THE
advertisement

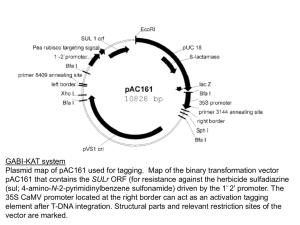
SEARCHING A FUNCTION FOR PPI1, AN INTERACTOR OF THE PLASMA MEMBRANE H+-ATPase, USING INSERTIONAL MUTANTS Piero Morandini1, Matteo Beretta Piccoli1, Giada Marino1, Chiara Consonni2, Ralph Panstruga2 and Carlo Soave1 1 Dept. of Biology, University of Milan. 2 Max Planck Institut fuer Zuechtungsforschung, Dept. of Plant Pathogen Interactions, Cologne, Germany PPI1 (PROTON PUMP INTERACTOR 1) is a protein identified in a two hybrid screen whose N-terminus binds to the Arabisopsis thaliana plasma membrane proton pump (PM H+-ATPase). The entire protein, or fragments thereof, purified as fusion protein from E. coli are able to stimulate the activity of the proton pump in purified membranes. To study the function of the PPI1 protein we are using a reverse genetic approach, i.e. identifying and characterizing Arabidopsis lines bearing a T-DNA inserted in the gene. Three T-DNA insertions were identified (and confirmed by sequencing of the amplified genomic DNA) in the available mutagenized collections (http://www.tmri.org or http://signal.salk.edu) for Ppi1: an insertion in intron I, one in exon VI at aa 525 and one in the promoter region (-235 from the transcription start site). For Ppi2, a gene with 52% similarity to Ppi1, we identified insertions at aa34 and 550 (also confirmed by sequencing). Homozygous plants for the various insertions were identified and back-crossed with the wt Col-0. A double mutant ppi1,ppi2 was also generated by crossing individual knockouts. RT-PCR analysis on single insertion lines confirmed the transcript is either absent or aberrant for insertions within both genes, while the insertion in the promoter of Ppi1 did not affect the mRNA structure, as seen by RT-PCR. Western blotting on homozygous knock-out plants are being performed to confirm that also the protein is aberrant or absent for the insertions within the gene. Both the single and the double mutant did not show evident phenotype when grown in pots. The single mutants have been exposed to several pathogens (powdery mildew from barley, wheat and Arabidopsis as well as Pseudomonas syringae), to wounding without showing differences in respect to wt plants, both on the macroscopic or microscopic level (staning with DAB or Trypan blue). Different growth conditions are being tested for identifying phenotypic differences to wt.