Functional redundancy and interaction in biological
advertisement
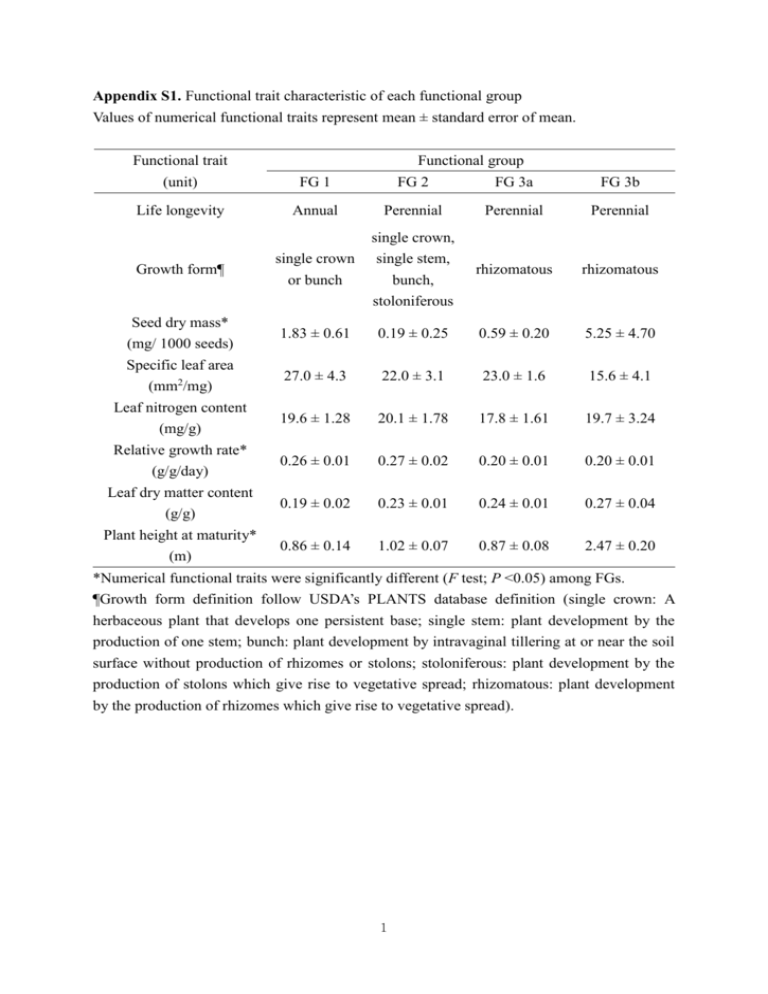
Appendix S1. Functional trait characteristic of each functional group Values of numerical functional traits represent mean ± standard error of mean. Functional group Functional trait (unit) FG 1 FG 2 FG 3a FG 3b Life longevity Annual Perennial Perennial Perennial single crown or bunch single crown, single stem, bunch, stoloniferous rhizomatous rhizomatous 1.83 ± 0.61 0.19 ± 0.25 0.59 ± 0.20 5.25 ± 4.70 27.0 ± 4.3 22.0 ± 3.1 23.0 ± 1.6 15.6 ± 4.1 19.6 ± 1.28 20.1 ± 1.78 17.8 ± 1.61 19.7 ± 3.24 0.26 ± 0.01 0.27 ± 0.02 0.20 ± 0.01 0.20 ± 0.01 0.19 ± 0.02 0.23 ± 0.01 0.24 ± 0.01 0.27 ± 0.04 0.86 ± 0.14 1.02 ± 0.07 0.87 ± 0.08 2.47 ± 0.20 Growth form¶ Seed dry mass* (mg/ 1000 seeds) Specific leaf area (mm2/mg) Leaf nitrogen content (mg/g) Relative growth rate* (g/g/day) Leaf dry matter content (g/g) Plant height at maturity* (m) *Numerical functional traits were significantly different (F test; P <0.05) among FGs. ¶Growth form definition follow USDA’s PLANTS database definition (single crown: A herbaceous plant that develops one persistent base; single stem: plant development by the production of one stem; bunch: plant development by intravaginal tillering at or near the soil surface without production of rhizomes or stolons; stoloniferous: plant development by the production of stolons which give rise to vegetative spread; rhizomatous: plant development by the production of rhizomes which give rise to vegetative spread). 1 Appendix S2. Overview of experiment (photo taken in summer 2009) 2 Appendix S3. Partitioning diversity effect Original equation about partitioning diversity effect (Loreau & Hector 2001) was modified by replacing response variable, yield with RCIavg (relative competitive effect on P. australis), as follows. Y Yo Ye RYo ,i M i RYe ,i M i RYi M i N RY M N cov( RYi , M i ) i i i Where, ∆Y = net diversity effect in term of relative competitive effect on P. australis (RCIavg) in a mixture Yo = observed competitive effect in the mixture Ye = expected competitive effect in the mixture RYo,i = Yo,i/ Mi = observed relative competitive effect of species i in the mixture Mi=competitive effect of species i in the monoculture Yo,i= Pi×Yo Pi = Relative cover of species i in the mixture (Note: it was originally relative y) RYe,i= proportion of species i in the mixture ∆RYi = RYo,I - RYe,i = deviation from expected relative competitive effect of species i in the mixture. In this equation, NRCM represents the complementarity effect, and N cov( RC i , Mi ) represents selection effect in terms of competitive effect. This equation is based on the assumption that invasion resistance of each species in mixture is proportional to “plant cover” of the species in the mixture (Co,i= Pi×Co). Because there was significant and positive relationship between plant cover of resident species and RCIavg in simple linear regression model (F1,55=71.96; P <0.001), the assumption was reasonable (Dr. Michel Loreau, personal communication). 3 Appendix S4. Changes in plant cover of wetland plants and P. australis from 2009 to 2010 August July Legend September Control 1 Group 1 Group 2 2010Plant cover of Phragmites australis2009 Plant cover of Phragmites australis Group 3 0.5 0 1 0.5 0 0 0.5 1 0 0.5 1 Plant cover of resident plants 0 0.5 Plant cover of wetland plants Legend Control □ FG 1 4 FG 2 × FG 3a 1 Appendix S5. Summary of the results of the diversity interaction models. Diversity interaction models (model term description) Model 1 (species identity effect) Model 2 (functional group identity No. of terms 11 3 AIC¶ Model terms (P <0.05) model term P Estimate βLolium βBidens -14.43 βMimulus βScirpus βCalamagrostis <0.001 <0.001 0.012 0.0474 0.0475 1.190 0.712 0.396 0.308 0.311 βFG1 -16.44 βFG2 <0.001 0.012 0.971 0.339 βFG1 βFG2 -26.16 δav <0.001 0.023 0.001 0.895 0.281 0.436 βFG1 βFG2 <0.001 0.004 <0.001 0.817 0.308 2.219 <0.001 <0.001 0.001 0.001 0.021 0.832 0.319 5.222 5.047 2.011 <0.001 0.006 <0.001 0.811 0.295 2.098 effect): Model 3 (functional group identity effect and average species interaction) Model 4 (functional group identity effect and species interaction within and between functional group) Model 5 (functional group identity effect and separate species interactions) 4 9 -43.12 δFG1 FG3 βFG1 βFG2 δLolium 58 -41.26 Panicum δLolium Mimulus δLolium Scirpus Model 6 (functional group identity effect and species interaction between 6 -44.46 functional group) ¶Akaike information criterion 5 βFG1 βFG2 δFG1 FG3 Appendix S6. Average similarity coefficient of wetland plants to P. australis in each functional group. Error bar shows standard error of mean. Functional groups connected by same letter are not significantly different from each other. australis Phragmitesaustralis coefficient totoPhragmites Similaritycoefficient Similarity Chart 0.9 0.8 A A 0.7 B 0.6 0.5 C 0.4 0.3 0.2 0.1 0 FG 11 FG 22 FG 3a3a FG 3b3b Functional FG group Each error bar is constructed using 1 standard error from the mean. 6 100.0 50 1.2 60 2.0 1.2 1.0 0.8 0.4 0.2 0.30 1.2 r= -0.36; P=0.006 0.0 0.2 0.4 0.6 RCI.avg 0.8 1.0 1.5 0.26 Relative Growth Rate (g/g/day) 0.4 1.0 0.22 1.0 1.2 Growth form 0.2 0.5 Seed mass (g/ 1000 seeds) r= 0.51; P <0.001 0.18 0.0 0.0 0.2 0.4 0.6 RCI.avg 0.8 1.0 (h) stoloniferous Rhizomatous 0.0 (f) 0.0 0.0 0.2 0.4 C single crown r= 0.59; P <0.001 150 0.6 1.2 1.0 0.8 RCI.avg BC Stoloniferous perennial 100 Plant cover (%) AB Bunch (g) 0.8 1.0 40 A Single crown 1.2 30 0.6 1.2 1.0 0.8 0.6 0.0 0.2 0.4 RCI.avg B (e) Life longevity RCI.avg 20 Plant height (cm) A annual 50 0.2 10 Biomass (g/pot) (d) 0 0.0 0 0.8 20.0 0.6 5.0 RCI.avg 1.0 r= 0.79; P <0.001 0.6 RCI.avg 0.8 0.6 RCI.avg 0.2 0.0 0.2 (c) 0.4 1.2 r= 0.61; P <0.001 0.4 0.8 0.6 0.4 0.0 0.2 RCI.avg (b) 1.0 (a) r= 0.77†; P <0.001‡ 1.0 1.2 Appendix S7. Relationship between plant traits and biotic resistance (RCIavg) in the first experiment. Performance trait (from experiment): (a) biomass, (b) plant height, and (c) plant cover; Functional traits (from TRY trait database): (d) life longevity, (e) growth form, (f) RGR, (g) seed mass, and (h) LDMC. Solid line represents linear regression fit (log-scale was used in case of biomass). Means connected by same letter are not significantly different from each other in ANOVA test (life longevity: F1,29=63.83; P<0.001; growth form: F3,27=10.81; P<0.001). Only functional traits that have significant relationship with RCIavg are shown. †r values represent Pearson correlation coefficient. ‡P values represent t test result on slope in linear regression analysis. 0.20 0.25 0.30 Leaf dry matter content (g/g) 7 0.35 0.6 0.8 1.0 1.2 1.4 Plant height at maturity (m) RCI.meanRCI.biomass RCI.shoot RCI.coverRCI.heightphrag.biomass RCI.mean 1.0000 0.9547 0.9688 0.9368 0.8028 -0.9293 RCI.biomass 0.9547 1.0000 0.9225 0.8456 0.7005 -0.9399 RCI.shoot 0.9688 0.9225 1.0000 0.8998 0.7141 -0.8797 RCI.cover 0.9368 0.8456 0.8998 1.0000 0.6490 -0.8859 RCI.height 0.8028 0.7005 0.7141 0.6490 1.0000 -0.6618 Appendix S8. Correlations among different response variables (RCIavg, RCInumber of shoot, phrag.biomass -0.9293 -0.9399 -0.8797 -0.8859 -0.6618 1.0000 RCIbiomass, RCIheight, RCIplant cover, and biomass of P. australis) in the experiments. correlations are estimated by REML method. †rThe values represent Pearson correlation coefficient. Scatterplot M atrix 1 0.5 RCIavg RCI.mean 0 r=0.95† -0.5 1 0.5 r=0.96 r=0.93 r=0.80 r=-0.92 r=0.92 r=0.84 r=0.70 r=-0.93 r=0.89 r=0.71 r=-0.87 r=-0.88 RCI.biomass RCIbiomass 0 -0.5 1 0.5 RCI.shoot RCIshoots 0 -0.5 1 0.6 RCI.cover RCIcover 0.2 r=0.64 -0.2 1 r=-0.66 0.6 RCI.height RCIheight 0.2 -0.2 70 50 30 10 -10 phrag.biomass -0.5 0.5 1 -0.5 0.5 1 -0.5 0.5 1 -0.2 0.4 1 -0.2 0.4 1 -10 30 60 Multivariate year=2010 Sept Correlations RCI.meanRCI.biomass RCI.shoot RCI.coverRCI.heightphrag.biomass RCI.mean 1.0000 0.9573 0.8731 0.8846 0.7843 -0.9586 RCI.biomass 0.9573 1.0000 0.7796 0.7899 0.7364 -0.9998 RCI.shoot 0.8731 0.7796 1.0000 0.7027 0.5569 -0.7819 RCI.cover 0.8846 0.7899 0.7027 1.0000 0.5923 -0.7903 RCI.height 0.7843 0.7364 0.5569 0.5923 1.0000 -0.7400 phrag.biomass -0.9586 -0.9998 -0.7819 -0.7903 -0.7400 1.0000 The correlations are estimated by REML method. Scatterplot M atrix 0.5 0.2 -0.1 RCI.mean 8 Appendix S9. Analysing biomass of P. australis in monoculture experiments In 2009 for the first experiment, biomass of P. australis was significantly different among three FGs (F2,20= 25.21 , P<0.001), but it was not significantly different within each FG (F8,20= 2.00, P=0.098). In 2010 for the first experiment, biomass of P. australis was significantly different among three FGs (F2,20= 28.14 , P<0.001), but it was not significantly different within each FG (F8,20= 0.96, P=0.492). In 2011 for the second experiment, biomass of P. australis was significantly different among four FGs (F3,48= 8.96, P<0.001), but it was not significantly different within each FG (F21,48= 1.73, P=0.059). 9 150 100 Selection effect Net diversity effect Complementarity effect experiment Net effect mixture Mixture 50 60 40 20 mono Monoculture (b) 0 Diveristy effecteffect (biomass of resident species) (Biomass) in mixture Diversity 100 120 P <0.001† 80 (a) 0 Biomass (g/pot) plants (g/pot) wetland species Biomassofofresident Appendix S10. Partitioning diversity effect based on biomass of resident species in mixture experiment. †Contrast test result Selection effect Partitioning diversity effect 10