The Q and T parameters were varied in an attempt to maximize the
advertisement
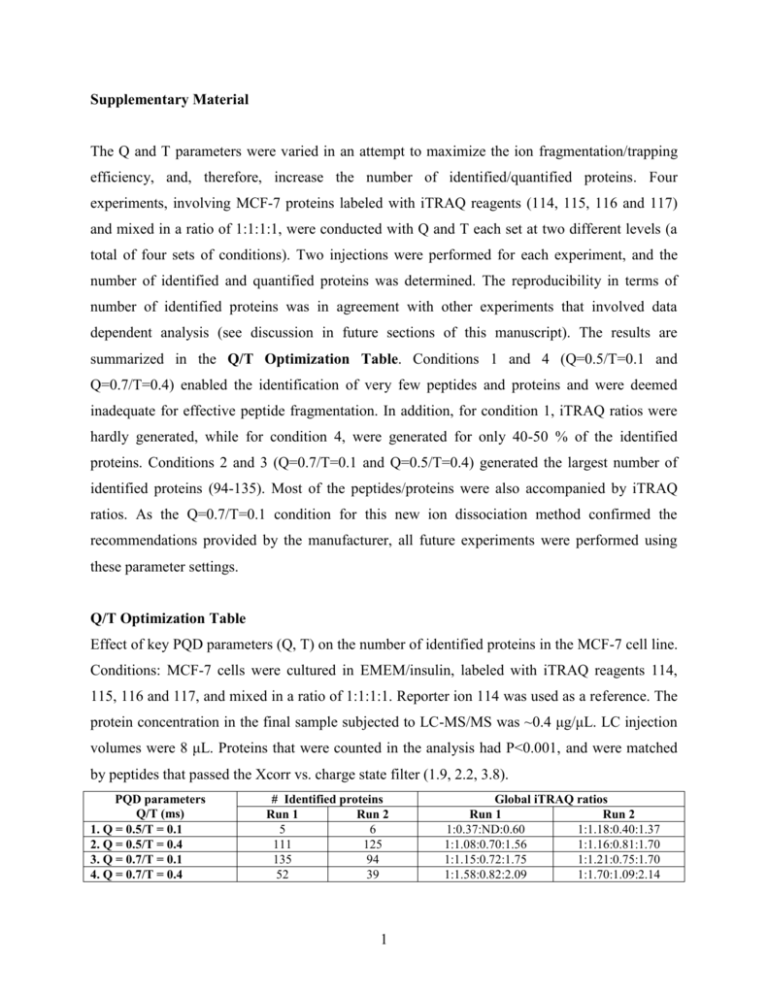
Supplementary Material The Q and T parameters were varied in an attempt to maximize the ion fragmentation/trapping efficiency, and, therefore, increase the number of identified/quantified proteins. Four experiments, involving MCF-7 proteins labeled with iTRAQ reagents (114, 115, 116 and 117) and mixed in a ratio of 1:1:1:1, were conducted with Q and T each set at two different levels (a total of four sets of conditions). Two injections were performed for each experiment, and the number of identified and quantified proteins was determined. The reproducibility in terms of number of identified proteins was in agreement with other experiments that involved data dependent analysis (see discussion in future sections of this manuscript). The results are summarized in the Q/T Optimization Table. Conditions 1 and 4 (Q=0.5/T=0.1 and Q=0.7/T=0.4) enabled the identification of very few peptides and proteins and were deemed inadequate for effective peptide fragmentation. In addition, for condition 1, iTRAQ ratios were hardly generated, while for condition 4, were generated for only 40-50 % of the identified proteins. Conditions 2 and 3 (Q=0.7/T=0.1 and Q=0.5/T=0.4) generated the largest number of identified proteins (94-135). Most of the peptides/proteins were also accompanied by iTRAQ ratios. As the Q=0.7/T=0.1 condition for this new ion dissociation method confirmed the recommendations provided by the manufacturer, all future experiments were performed using these parameter settings. Q/T Optimization Table Effect of key PQD parameters (Q, T) on the number of identified proteins in the MCF-7 cell line. Conditions: MCF-7 cells were cultured in EMEM/insulin, labeled with iTRAQ reagents 114, 115, 116 and 117, and mixed in a ratio of 1:1:1:1. Reporter ion 114 was used as a reference. The protein concentration in the final sample subjected to LC-MS/MS was ~0.4 μg/μL. LC injection volumes were 8 μL. Proteins that were counted in the analysis had P<0.001, and were matched by peptides that passed the Xcorr vs. charge state filter (1.9, 2.2, 3.8). PQD parameters Q/T (ms) 1. Q = 0.5/T = 0.1 2. Q = 0.5/T = 0.4 3. Q = 0.7/T = 0.1 4. Q = 0.7/T = 0.4 # Identified proteins Run 1 Run 2 5 6 111 125 135 94 52 39 1 Global iTRAQ ratios Run 1 Run 2 1:0.37:ND:0.60 1:1.18:0.40:1.37 1:1.08:0.70:1.56 1:1.16:0.81:1.70 1:1.15:0.72:1.75 1:1.21:0.75:1.70 1:1.58:0.82:2.09 1:1.70:1.09:2.14 Collision Energy Settings We present below the results of an experiment that involved the analysis of two cell states of MCF-7 cells. Two replicates of each cell state were labeled with two iTRAQ reagents each (cell state 1 with reagents 114 and 115, and cell state 2 with reagents 116 and 117) and mixed in a ratio of (1:1):(1:1). The sample was analyzed by LC-MS/MS using the conditions described in the experimental section and CE settings of 23 %, 28 %, 31 % and 35 %. All other conditions were kept constant. In this particular experiment, the number of protein I.D.s max out at CE=31 %, while at CE=23 %, iTRAQ ratios were not measurable. The number of identified proteins with P<0.001 and the global correction factors for each cell state are summarized in the CE Setting Table. The optimal CE values changed in time, thus frequent optimization is probably highly advisable. CE Setting Table CE setting, % # Identified proteins Global iTRAQ ratios 23 10 iTRAQ ratios could not be generated 28 85 1 : 1.06 : 0.68 : 0.53 31 87 1 : 0.99 : 0.63 : 0.50 35 58 1 : 1.06 :0.73 : 0.72 2 Scatter Plot of Average Z-scores for the 255 Protein Set Conditions: MCF-7 cells were grown in the presence of E2 and Tam using the following conditions: (A) E2 (1 nM) and (C) E2 (10 pM)/Tam (1 μM). Two experimental replicates were performed for each cell state. Protein digests from cell condition A were labeled with reagents 114 and 115, while protein digests from cell condition C were labeled with reagents 116 and 117. The samples were mixed in a ratio (A:A):(C:C) of (1:1):(1:1). The protein amount consumed for each labeling reaction was 100 μg. Multiconsensus data from five LC-MS/MS experiments were used in the analysis (8 μL injections, ~2 μg/μL protein concentration). Peptide level filtering was performed with the Xcorr vs. charge state filter set at 1.5, 2.0, 3.0. Protein (P<0.001) level filtering is described in the text. Average Z-score 6.00 4.00 2.00 0.00 -2.00 -4.00 -6.00 0 25 50 75 100 125 150 175 200 225 250 275 Protein # 3 Tandem Mass Spectrum of a Relevant Peptide Belonging to Poly(rC) Binding Protein 1. The inset represents an enlarged view of the iTRAQ reporter ions m/z region. #6405-6405 NL: 1.20E2 +1 y4 616.5 100 Poly(rC)-binding protein 1 95 Q*QSHFAMMHGGTGFAGIDSSSPEVK* (3+) MH+=2894.38, -10lgP=87, Xcorr=4.85, Cn=0.43 90 85 75 #6405-6405 RT:92.34-92.34 NL: 1.20E2 70 65 +1 y1 291.2 60 y2 +2 390.4 b 6 422.4 50 +2 bo 12 720.7 45 +1 40 +3 a 17 +2 y* 11 608.4 +1 bo 4 607.5 +2 y9 553.3 30 25 +2 y* 6 386.8 20 +3 10 +1 b6 y7 877.5 625.4 35 +1 843.5 b5 +1 772.5 y5 703.5 90 85 80 75 116 70 65 60 55 115 50 45 40 35 30 117 114 25 20 15 10 5 0 110 111 112 113 114 115 116 117 118 m/z 119 120 121 122 123 124 125 +1 m/z +1 +2 +1 +1 95 b7 974.7 y* 18 952.8 y3 b1 174.2 273.2 a1 245.3 100 Relative Abundance +1 55 15 Relative Abundance Relative Abundance Relative Abundance 80 y* 8 975.5 +1 y9 +1 b8 +2 1105.7 1105.7 +1 y 10 a 24 1288.2 1162.6 +3 b7 325.5 +1 +2 +1 b 20 1096.9 5 +1 y 13 b 13 1438.0 1514.8 y* 16 1636.1 0 200 400 600 800 1000 m/z m/z 4 1200 1400 1600 1800 126










