Supplementary Material
advertisement

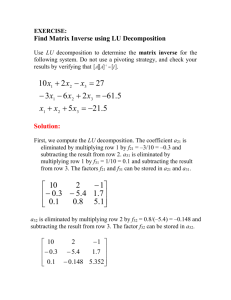
Supplementary Materials Supplementary Tables Supplementary Table S1. ChIP-Seq Datasets [1] Dataset DM230 Mark PolII (RNA polymerase II) Species/Tissue Mouse/Liver GEO Accession GSM722763 DM05 p300 (co-activator protein) Mouse/Liver GSM722762 DM721 H3K27ac (H3 lysine 27 acetylation) Mouse/Liver GSM851275 Supplementary Table S2. Motif Datasets [1] Case Motif Dataset Motif Input Format Study 1 2 3 Number of ChIP-Seq Motifs Dataset DREME_DM230 MEME’s output 1 DM230 MEME_DM230 MEME’s output 20 DM230 PScanChIP_DM230 Jaspar 14 DM230 RSAT_peak-motifs_DM230 TRANSFAC-like 10 DM230 W-ChIPMotifs_DM230 PSSM 11 DM230 MEME_DM05 MEME’s output 46 DM05 MEME-ChIP_DM05 MEME’s output 4 DM05 PScanChIP_DM05 Jaspar 16 DM05 RSAT_peak-motifs_DM05 TRANSFAC-like 17 DM05 W-ChIPMotifs_DM05 PSSM 11 DM05 DREME_DM721 MEME’s output 16 DM721 MEME-ChIP_DM721 MEME’s output 11 DM721 PScanChIP_DM721 Jaspar 37 DM721 RSAT_peak-motifs_DM721 TRANSFAC-like 40 DM721 1 Supplementary Figures Supplementary Figure S1: An illustration of a motif in a position specific probability matrix used by MOTIFSIM. The four columns represent A, C, G, and T nucleotides respectively. The length of this motif is seven, which is the number of rows in the matrix. Each element in the matrix is a probability value of a nucleotide. Supplementary Figure S2: Network architecture of MOTIFSIM web tool. The HAProxy load balancer directs web traffic to different Apache web server nodes on the cluster. 2 Conversion of different motif input formats to position specific probability matrices The procedures for converting different motif input formats, which are described in the user manuals, to position specific probability matrices [2] used by MOTIFSIM are slightly different because of their different structures. An illustration of a position specific probability matrix used by MOTIFSIM is in Figure S1. 1. TRANFAC, TRANFAC-like, Position Specific Scoring Matrix (PSSM), and vertical matrix formats to position specific probability matrices We applied the same conversion procedure for these motif input formats because they have four columns for A, C, G, and T nucleotides. The sum of four values for A, C, G, and T in each row for each input format must be identical. To convert the input matrix (green matrix below) to a position specific probability matrix (yellow matrix below), each element in a position specific probability matrix is calculated by taking the value of each element in the input matrix divided by the sum of the four values in each row in the input matrix. The sum of the four values for A, C, G, and T in each row of a position specific probability matrix must be 1. A 73 44 485 0 79 0 C 81 578 65 570 0 0 G 407 0 0 52 0 622 T 61 0 72 0 543 0 Sum 622 622 622 622 622 622 A 0.1174 0.0707 0.7797 0.0000 0.1270 0.0000 C 0.1302 0.9293 0.1045 0.9164 0.0000 0.0000 G 0.6543 0.0000 0.0000 0.0836 0.0000 1.0000 T 0.0981 0.0000 0.1158 0.0000 0.8730 0.0000 2. Jaspar and horizontal matrix formats to position specific probability matrices 3 Sum 1 1 1 1 1 1 The Jaspar and horizontal matrix formats have four rows represent A, C, G, and T nucleotides respectively. The sum of four values in each column must be identical. To convert the input matrix (blue matrix below) to a position specific probability matrix (yellow matrix below), we divide the value of each element in each column of the input matrix by the sum of four values in that same column. The results of the entire column are transposed to make the entire row for the position specific probability matrix. A C G T [ [ [ [ Sum 8 1 3 2 13 0 1 0 0 0 13 1 3 0 11 0 2 2 0 10 0 13 0 1 14 0 0 0 3 8 2 1 14 14 14 14 14 14 14 14 ] ] ] ] A 0.5714 0.9286 0.0000 0.2143 C 0.0714 0.0000 0.0000 0.0000 G 0.2143 0.0714 0.9286 0.7857 T Sum 0.1429 1 0.0000 1 0.0714 1 0.0000 1 0.1429 0.0000 1.0000 0.2143 0.1429 0.9286 0.0000 0.5714 0.0000 0.0000 0.0000 0.1429 0.7143 0.0714 0.0000 0.0714 1 1 1 1 3. Consensus sequence format to position specific probability matrices Each letter in a consensus sequence is converted to one row of a position specific probability matrix using the IUPAC nucleotide code [3] in the table below. IUPAC Nucleotide Code Base A C G T (or U) R Y S W K M B D Adenine Cytosine Guanine Thymine (or Uracil) A or G C or T G or C A or T G or T A or C C or G or T A or G or T Corresponding Row in Position Specific Probability Matrix 1.0000 0.0000 0.0000 0.0000 0.0000 1.0000 0.0000 0.0000 0.0000 0.0000 1.0000 0.0000 0.0000 0.0000 0.0000 1.0000 0.5000 0.0000 0.5000 0.0000 0.0000 0.5000 0.0000 0.5000 0.0000 0.5000 0.5000 0.0000 0.5000 0.0000 0.0000 0.5000 0.0000 0.0000 0.5000 0.5000 0.5000 0.5000 0.0000 0.0000 0.0000 0.3333 0.3333 0.3333 0.3333 0.0000 0.3333 0.3333 4 H V A or C or T A or C or G N any base 0.3333 0.3333 0.2500 0.3333 0.3333 0.2500 0.0000 0.3333 0.2500 0.3333 0.0000 0.2500 4. Sequence alignment format to position specific probability matrix First, the sequence alignment is converted to a profile matrix, which has four rows representing A, C, G, and T nucleotides. Then, the profile matrix is converted to a position specific probability matrix. An example of this conversion is below. A C G T Sum AACACGTGGC A 3 3 2 4 0 0 0 0 0 1 0.6000 0.2000 0.2000 0.0000 1 GCCACGTGCC C 1 1 3 1 4 1 0 0 2 2 0.6000 0.2000 0.2000 0.0000 1 CGCATGTGCA G 1 1 0 0 0 4 0 4 1 1 0.4000 0.6000 0.0000 0.0000 1 AAAACGTGTT T 0 0 0 0 1 0 5 1 2 1 0.8000 0.2000 0.0000 0.0000 1 AAACCCTTTG Sum 5 5 5 5 5 5 5 5 5 5 0.0000 0.8000 0.0000 0.2000 1 0.0000 0.2000 0.8000 0.0000 1 0.0000 0.0000 0.0000 1.0000 1 0.0000 0.0000 0.8000 0.2000 1 0.0000 0.4000 0.2000 0.4000 1 0.2000 0.4000 0.2000 0.2000 1 Sequence Alignment Profile Matrix Position Specific Probability Matrix Calculate the absolute value of the difference between each pair of corresponding matrix elements The difference between two corresponding matrix elements p i j ,k and p i 1 j , k in the overlapping window p p between two matrices pi and pi 1 is calculated by subtracting element i j , k from element i 1 j , k . The difference can be positive or negative. Therefore, the absolute value of the difference is calculated. The example below illustrates this process. The bold rectangle is the overlapping window. The difference 5 between the first element (red) in matrix pi (yellow) and the first element (red) in matrix pi 1 (green) is calculated in the first element (red) of the deference matrix d i , i 1 (purple). References 1. Tran, N.T.L. and C-H. Huang. 2014. A survey of motif finding Web tools for detecting binding site motifs in ChIP-Seq data. Biology Direct 9:1-22. 2. Li, H. 2002. Computational approaches to identifying transcription factor binding sites in yeast genome. Methods in Enzymology: Guides to Yeast Genetics and Molecular Biology, Part B 350:484-495. 3. IUPAC nucleotide code. Available at http://www.bioinformatics.org/sms/iupac.html. 6