BIOTECH Laboratory Activity:
advertisement

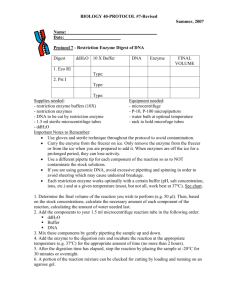
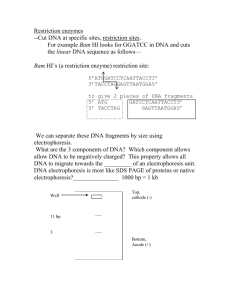
Biotechnology in Medicine and Agriculture: Analysis of Transformation, Plasmid Prep and Restriction Digest I. Objectives Upon completion of this laboratory, you should be able to: Extract plasmid DNA from E. coli cells Ethanol precipitate DNA from aqueous solutions Explain how restriction enzymes work and where they come from Set up restriction enzyme digests of DNA II. Safety Considerations As described in the last lab, treat E. coli as biohazardous waste while they are alive. NaOH/SDS and KOAc are both fairly strong irritants, so handle them carefully. In particular, if you get either in your eyes, flush well with water. III. Introduction RESTRICTION ENZYMES Viruses called bacteriophages are major enemies of bacteria. These viruses infect bacteria by injecting their own DNA into bacteria to force the bacteria to replicate their DNA and to synthesize the protein coats that enclose the DNA (or RNA) of a mature virus. In response to this strong selection pressure, a natural defense system has evolved in bacteria. A major component of this system are restriction enzymes that cut up and destroy the invading DNA. (These enzymes are most accurately referred to as endonucleases since they attack DNA at internal sites in the molecule rather than chewing it up from the ends –enzymes that do that are called exonucleases.) A restriction enzyme acts like molecular scissors, making cuts at specific sequences of base pairs that it recognizes. In doing this, they bind DNA and slide along the double helix until specific sequences of base pairs are recognized - signaling the enzyme to stop sliding. The enzyme then cuts or chemically separates the DNA molecule at that site, which is called a restriction site. Bacteria prevent digestion of their own DNA by chemically modifying certain DNA bases within the specific enzyme recognition sequence (usually by adding a methyl group to one of the 1 nitrogenous bases), which prevents the restriction enzymes from recognizing their sites. This could be considered a very primitive immune system. Restriction enzymes typically recognize palindromic sequences of base pairs - sequences that have the same sequence when read 5' to 3' on each strand. An example of a 6 base-pair palindromic sequence is given below. 5'-GAATTC-3' 3'-CTTAAG-5' Some restriction enzymes may leave short unpaired nucleotide bases, called “sticky” ends, at the DNA sites where they cut, whereas other restriction enzymes make a cut in the middle, creating double stranded DNA fragments with “blunt” ends. Below is an example of a "sticky cut." Note that when the DNA molecule is cut in this way, the ends of the resulting molecules have unpaired nucleotides. 5'-G A A T T C-3' 3'-C T T A A G-5' 5'-G-3' 3'-C T T A A -5' 5'-A A T T C-3' 3'-G-5' Example of a restriction enzyme cut resulting in "sticky," unpaired ends. In a blunt cut, there are no unpaired nucleotides because the enzyme cuts its restriction site right in the middle. 5'-G A A T T C-3' 3'-C T T A A G-5' 5'-G A A -3' 3'-C T T -5' 5'- T T C-3' 3'- A A G-5' Example of a restriction enzyme cut resulting in "blunt" ends. If a specific restriction site occurs in more than one location on a DNA molecule, a restriction enzyme will make a cut at each of those sites, resulting in multiple fragments. Therefore, if a given linear piece of DNA is cut with a restriction enzyme whose specific recognition code is 2 found at two different locations on the DNA molecule, the result will be three fragments of different lengths. If the given piece of DNA is circular and is cut with a restriction enzyme whose specific recognition code is found at two different locations on the DNA molecule, the result will be two fragments of different lengths. The length of each fragment will depend upon the location of restriction sites on the DNA molecule. One common restriction enzyme is Eco RI. This enzyme was isolated from E. coli (hence the "Eco" in its name) and has the 6 base-pair recognition site 5'-GAATTC-3'. It cuts its sequence as shown below. Consider the two samples of DNA shown below. Sample #1 5' - C A G T G A T C T C G A A T T C G C T A G T A A C G T T - 3' 3' - G T C A C T A G A G C T T A A G C G A T C A T T G C A A - 5' Sample #2 5' - T C A T G A A T T C C T G G A A T T C G C A A A T G C A - 3' 3' - A G T A C T T A A G G A C C T T A A G C G T T T A C G T - 5' Pre-lab Question: If both samples are treated with the Eco R1 restriction enzyme, how many fragments would be produced? Circle the recognition sites of the enzyme. Like all enzymes, restriction enzymes function best under specific buffer and temperature conditions. Eco RI functions best at 37 o C and, when cutting DNA in vitro (outside of the cell in a test tube), it prefers a reaction buffer that consists of various salts and other reagents that hold the pH at 8.0. Pre-lab Question: Why do you think the temperature optimum for EcoRI is 37 oC? III. Procedure A. Analyzing transformation results 1. Record the number of colonies on each of your plates. If there are too many to count, try to count a representative group (such as a quadrant) and extrapolate or estimate the number of colonies on the entire plate. Compare and contrast the growth on each of the plates. 3 2. Use long wave UV light to detect GFP expression in the bacteria on each plate and record the approximate number and percentage of colonies expressing GFP on each plate. B. pGLO Plasmid DNA extraction from E. coli 1. A single, well-isolated colony from one or more of the LB+amp+pGLO+arabinose plates was inoculated into medium the night before your lab. It was allowed to grow overnight for you to extract the plasmid DNA today. 2. You will receive a bacterial overnight culture from your instructor. Label two 1.5 mL centrifuge tubes (Tube 1 and Tube 2). Begin by aliquoting 1 mL of your culture into your two 1.5 mL microcentrifuge tubes. Close the caps and spin them in the microcentrifuge for 1 min at the highest speed (14,000 rpm). Note – make sure to balance the centrifuge with your two tubes and make sure you have labeled them with your Group/Table name. 3. After the 1 min spin, you should have a visible pellet of cells at the bottom of each of your tubes. Without disturbing the pellet, pour off the supernatant in your liquid waste. Repeat steps 2 and 3 using the same set of tubes (this step is included to double the number of cells in each tube). 4. Add 100 uL of ice-cold glucose/Tris/EDTA solution (GTE) to each tube. Using the vortex machine at your lab bench, you will resuspend your pellets by firmly placing the tip of your tube into the cup of the vortex. Be sure that the “touch on” setting is used. Vortex each of your microcentrifuge tubes for approximately 30 seconds or until no visible clumps remain. 5. Add 200 uL of fresh NaOH/SDS cell lysis solution. Close the cap and mix your solution by gently inverting your tubes 5 times. Incubate your tubes on ice for 5 min. (The cells are killed by this step so material after this is not considered biohazard waste.) 6. Add 150 uL of ice-cold potassium acetate (KOAc) solution (precipitates chromatin, but not plasmid). Close the caps, and mix the solution by carefully inverting your tubes 5 times. A white precipitate should appear. Incubate on ice for 5 min. 7. Microcentrifuge for 5 min at full speed, 14,000 rpm. Transfer 400 µL of the supernatant from each tube into two new 1.5 mL microcentrifuge tubes (label each of the new tubes). Be sure to avoid pipeting the white precipitate and wipe off any precipitate clinging to outside of tip prior to expelling the supernatant. Discard old tubes containing precipitate. (If you do transfer some precipitate accidentally, tubes can be respun and supernatant removed to a fresh tube time following this spin.) 8. Add 400 µL of isopropanol to the supernatant tubes. Close the caps, and mix your solutions by gently inverting the tubes 5 times. Let stand at room temperature for only 2 min (proteins may precipitate with the DNA if allowed to incubate longer). 8. Microcentrifuge for 5 min at 14,000 rpm. Align your tubes in rotor so that cap hinge points outward AND remember to balance the centrifuge. The nucleic acid residue, visible or not, will collect on the tube side under the hinge during centrifugation. 4 9. Pour off the supernatant from the tubes (be careful not to disturb the nucleic acid pellet). Invert the tube and tap gently on surface of a clean paper towel to drain thoroughly. 10. Add 200 µL of 100% ethanol and close your tubes. Flick tubes several times to wash pellets. 11. Centrifuge the tubes at full speed (14,000 rpm) for 5 min. Pour off supernatant from the tubes (be careful not to disturb the nucleic acid pellet). Invert the tube and tap gently on surface of a clean paper towel to drain thoroughly. 12. Keep the lid off your tube and place it in the rack provided to allow your pellets to air-dry. If after 10 minutes the remaining ethanol has not evaporated, see your instructor regarding the application of a hair dryer. If this is indicated, be careful not to dislodge the pellets from the tubes! Place your tubes in a micro-tube plastic rack and apply a stream of warm air on the “low” setting for 3-5 minutes or until all visible ethanol has been evaporated. Be sure to hold the nozzle of the hair dryer no closer than 12-inches away from the mouths of the tubes. 13. When there is no liquid visible in your tubes, add 30 µL of TE buffer (Tris-EDTA) to each tube to dissolve your pellets. If the pellets do not resuspend easily (by flicking the bottom of the tube), close the caps and use a P200 pipet to gently resuspend your pellets. 14. You should now have two 30 L tubes (Tube 1 and Tube 2). This is your plasmid DNA that you will digest using restriction enzymes in part C. C. Restriction enzyme analysis of plasmid DNA You are going to set up two restriction digests using two different restriction enzymes: EcoRI and HindIII. You will perform this process on the DNA from Tubes 1 and 2 from your plasmid prep, so each lab group will have a total of 4 restriction digests. You will first need to obtain 4 new 1.5 mL centrifuge tubes. It is recommended that you label your new tubes as shown here: Tube 1 A –digest with EcoRI Tube 1 B –digest with HindIII Tube 2 A –digest with EcoRI Tube 2 B –digest with HindIII The following reagents are necessary for a restriction digest: DNA sample Restriction enzyme buffer (10X) Restriction enzyme(s) Sterile water Some of the reagents have been prepared and combined for you in advance, so you will set up your digests according to the following instructions. (Note that a summary table is included below that you may find helpful.) 1. In Tube 1 A, add 10 µL of your plasmid prep DNA (from Tube 1) to 9 µL of EcoRI reaction buffer (your reaction buffer contains the 10X restriction enzyme buffer, BSA and water). You will repeat this step for Tube 2 A. DO NOT throw away the remaining DNA in Tube 1; you will use this in a future lab. 5 2. In Tube 1 B, add 10 µL of your plasmid prep DNA to 9 µL of HindIII reaction buffer. You will repeat this step for Tube 2 B. Tubes 1 and 2 B are for your HindIII digest. DO NOT throw away the remaining DNA in Tube 2; you will use this in a future lab. 3. The restriction enzymes will be available at the front of the lab – see your instructor when you are ready to add them. You will add 1 µL of EcoRI to each EcoRI digest and 1 µL of HindIII to each of your HindIII digests. Mix the enzymes with the rest of your reagents by flicking your tube, and put on ice until you are ready to incubate (step 5.). 4. If you introduced bubbles or splashed the reactions up to the lid or into droplets on the walls of the tube, a quick spin in the centrifuge will collect the sample at the bottom of the tube. 5. Place your tubes in the 37 o C water bath for incubation. Your tubes will be incubated for 2 hours to ensure that the digestion goes to completion. The instructor or TA will remove your tubes for you and put them in the refrigerator until the next lab meeting. 6. When you are done, bring your tube of plasmid DNA (Tubes 1 and 2) to the front bench for storage until the next lab. All other leftover reagents from the digestion can be put in the trash. Summary Table Tube # 1A 2A 1B 2B DNA 10 µl DNA A 10 µl DNA B 10 µl DNA A 10 µl DNA B EcoRI Buffer 9 µl 9 µl ----- HindIII Buffer ----9 µl 9 µl EcoRI 1 µl 1 µl ----- HindIII ----1 µl 1 µl Total Volume 20 µl 20 µl 20 µl 20 µl You will analyze the results of your restriction digests by gel electrophoresis in the next lab activity. Questions: Answer in your lab notebook Examine the restriction map provided in the introduction to your lab to answer the following questions. 1. Explain why EcoRI should cut your plasmid DNA only once, while HindIII should cut your plasmid DNA twice. 2. How many fragments of DNA do you expect to get from the EcoRI digestion? What size do you think this/these fragment(s) will be? 3. How many fragments of DNA do you expect to get from the HindIII digestion? What size do you think this/these fragment(s) will be? 6