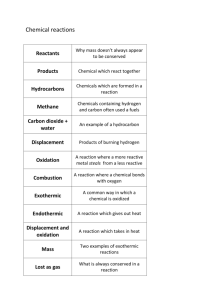
This course provides an introduction to bioinformatics, the combined
advertisement

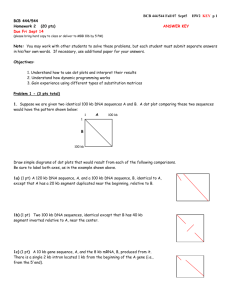
BIO 224 Laboratory CSU, Sacramento Dr. Tom Peavy February 13, 15 & 20, 2007 Assignment 3 (20 pts) (due Thurs Feb 22nd at 5pm) 1. Perform a blastp search using a highly conserved human protein as a query (integrin receptor beta 1, NP_391988.1). Use the default parameters, (nr or non-redundant database, search “All organisms”, and the BLOSUM62 matrix). In the formatting section, chose to view 500 Descriptions, Alignments and Graphic Overviews (scroll down in the format section and change the drop down numbers). A) Scroll down to the last page of the BLAST results and copy and paste the database info into this document (section begins after the sequences are presented). B) What was the effective length of the query sequence? How many sequences did the database examine? What was the effective length of the database? What do these pieces of information mean? C) How many database hits occurred in this search? How many HSP’s were successfully gapped? What does this mean? D) What was the threshold value used in this search? What does this mean? E) Examining the graphical and alignment displays on the first page of the BLAST results, what species and protein had the highest score and E-value (careful not to quote a hit for the same human protein sequence)? 2. Perform a similar blastp search using the human integrin sequence using the nr database (non-redundant), but this time search only the “Arthropoda”. A) Answer the following questions: i) How many hits were there. ii) What species and protein has the highest score and E-value? (record into table found in question C) iii) Were there any conserved domains? (examine the red link for conserved domain hits within the format section and follow the link). If so, describe the domain(s) that was found? B) Using the above Arthropoda BLAST search, at what score and E value do you suspect that the alignment is not for a homologous protein (meaning a non integrinrelated protein)? Provide your reasoning. C) Next fill in the table by repeating the search (same query, same nr database, same limitation to Arthopoda) using the two additional indicated scoring matrices. total # hits best score best E value first search, BL62 BLOSUM45 BLOSUM80 Was the same protein identified as the most closely related sequence in each of the searches? BIO 224 Laboratory CSU, Sacramento Dr. Tom Peavy February 13, 15 & 20, 2007 E) What was the effect of changing the scoring matrix with respect to the total number or hits, best score, and their E values. Explain why? (hint: think about the relationship of the scoring matrices in terms of matches-- which matrices give the highest scores for exact matches and highly conserved substitutions?) 3. Repeat above problem using a human protein that is poorly conserved, lactalbumin (NP_002280). Restrict the database to RefSeq and search “All organisms” using the default BLOSUM62 matrix. A) Answer the following: i) How many hits are there. ii) What species and protein has the highest score and E-value? (careful not to quote a hit for the same human protein sequence)? iii) Were there any conserved domains? (examine the red link for conserved domain hits within the format section and follow the link). If so, describe the domain(s) that was found? B) At what point can you discern that you are no longer examining the orthologous protein in your BLAST search results? Explain. C) Perform a similar blastp search using the human lactalbumin sequence using the nr database, but this time search only the “Arthropoda”. Fill in the chart below: Arthropoda total # matches best score best E value BLOSUM62 BLOSUM45 BLOSUM80 i) Were the same proteins identified as the best score/E value in all three searches? ii) Were the alignment over the whole length for the highest hits or only to portions of the lactalbumin protein? iii) What was the effect of changing the scoring matrix with respect to the total number or hits, best score, and their E values. Explain why? 4. What different search strategies might you use when studying a highly conserved protein (problem 2) versus a poorly conserved protein (problem 3)? (think about the various matrices and databases)