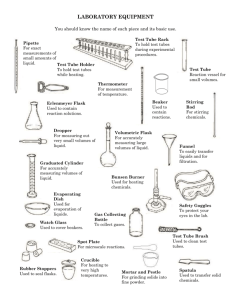
(Still Culture) ()
advertisement

CTAB Prep for Magnaporthe grisea (Still Culture)
1. Start a culture of the isolate in 30ml of CM media in a 100ml flask with an agar plug
of actively growing Magnaporthe or a clipping from a storage filter disk
(Penicillin/Streptomycin) can be added (to 100 ug/ml) to media to avoid problems with
bacterial contamination).
-Culture for 7-10 days in the dark until the mycelial mat in the flask has nearly filled the
flask bottom (regularly swirl flasks to avoid conidiation).
2. Remove the mycelial mat from flask with flamed forceps and blot dry with paper
towels, removing as much liquid as possible from the mat .
- proceed to step 3 or wrap in foil and freeze in liquid nitrogen and store at -70oC.
3. Place the sample in a mortar with liquid nitrogen and quickly grind the sample to a
fine powder (mycelia should powder easily).
4. Transfer powder to an autoclaved Sarstedt tube containing 4mL 2X CTAB buffer at
65C cap tubes and incubate for 20 min@65o, inverting the tube every 10 minutes to
further break up fungal material (this step inactivates cellular proteins and allows the
CTAB to complex with contaminating carbohydrates).
5. Add 4 ml CIA {Chloroform/pentanol (24:1)} to the tubes and let stand uncapped in
the fume hood for 1 minute (otherwise the caps may pop off the tubes).
cap tubes and gently mix on shaking platform for 20 minutes.
centrifuge 10 min@10 K @4oC (in fixed angle or swinging bucket rotor). REMOVE
LIDS FROM TUBES BEFORE CENTRIFUGING.
6. Carefully remove aqueous (top) phase (be sure not to take any of the organic phase or
cellular debris) and repeat Chloroform/pentanol extraction.
7. After the second Chloroform/pentanol wash, remove aqueous phase and add to 4mL
isopropanol in a Sarstedt tube, to precipitate nucleic acids.
at this step it is even more important not to pick up any of the CIA phase, so be very
careful to leave behind a few mm of the upper phase.
invert tube gently a few times to mix the sample and let sit for 5 minutes on ice to allow
complete precipitation of the nucleic acids (sometimes you can see the DNA at this point.
If you can't, don't panic).
8. Centrifuge at 10K/10 min./4oC in fixed angle rotor
Decant supernatant and drain tubes on a paper towel for about 15 min (at this point there
should be a white pellet at the bottom of the tube).
9. Resuspend the pellet in 500 ul 1X TE.
after the pellet is no longer visible transfer the sample to an eppendorf tube.
Phenol/chloroform (1:1) extraction. Reprecipitate the nucleic acids with 3 M NaOAc
(1/10th vol.) and 2 vol. of ethanol.
incubate at -20oC for 10 minutes and centrifuge at 13,000 rpm in microfuge for 20 min.
wash pellets once with 70% ethanol and spin in microfuge for 5 min.
drain and dry pellets on paper towel.
10. Resuspend pellets in 25-100 ul of TE+RNase (less if the pellets are small).
note: clean DNA pellets will resuspend completely at room temp. in only a few hours (do
not pipette solution up & down as this will tend to shear the DNA).
-samples may appear to be cloudy when resuspended, but this does not seem to interfere
with restriction digestion.
CTAB Extraction Solutions
CTAB Buffer [500ml]
CTAB
2%
10g
Trisma Base
100mM
6.06g
EDTA
10mM
1.46g
NaCl
0.7M
20.5g
note: CTAB is Hexadecyltrimethylammonium Bromide (Sigma # H-5882)
CIA
24 : 1 (V/V) Chloroform : Pentanol