1. introduction - NDSU Computer Science
advertisement

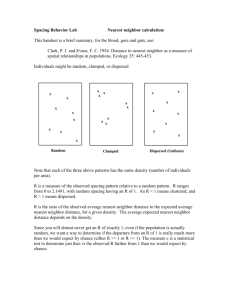
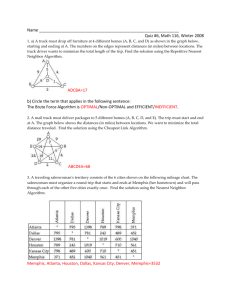
The Universality of Nearest Neighbor Sets in Classification and Prediction William Perrizo Gregory Wettstein, Amal Shehan Perera, Tingda Lu Computer Science Department North Dakota State University Fargo, ND 58105 USA Computer Science Department North Dakota State University Fargo, ND 58105 USA William.perrizo@ndsu.edu attributes (called the feature attributes) in a table (called the training table). ABSTRACT In this paper, we make the case that essentially all classification and prediction algorithms are nearest neighbor vote classification and predictions. The only issue is how one defines “near”. This is important because the first decision that needs to be made when faced with a classification or prediction problem is to decide which classification or prediction algorithm to employ. We will show how, e.g., Decision Tree Induction, as a classification method can be viewed legitimately as a particular type of Nearest Neighbor Classification. We will show how Neural Network Predictors are really Nearest Neighbor Classifiers. Keywords Predicate-Tree, Classification Nearest-Neighbor, Decision Tree Induction, Neural Network. 1. INTRODUCTION What is Data mining? Data mining, in its most restricted form can be broken down into 3 general methodologies for extracting information and knowledge from data. These methodologies are Rule Mining, Classification and Clustering. To have a unified context in which to discuss these three methodologies, let us assume that the “data” is in one relations, R(A1,…,An) (a universal relation – unnormalized) which can be thought of as a subset of the n product of the attribute domains, D i i 1 Rule Mining is a matter of discovering strong antecedentconsequent relationships among the subsets of the columns (in the schema). Classification is a matter of discovering signatures for the individual values in a specified column or attribute (called the class label attribute), from values of the other Clustering is a matter of using some notion of tuple similarity to group together training table rows so that within a group (a cluster) there is high similarity and across groups there is low similarity [20]. 1.1 P-tree Data Structure We convert input data to vertical Predicate-trees or Ptrees. P-trees are a lossless, compressed, and data-miningready vertical data structures. P-trees are used for fast computation of counts and for masking specific phenomena. This vertical data representation consists of set structures representing the data column-by-column rather than row-by row (horizontal relational data). Predicate-trees are one choice of vertical data representation, which can be used for data mining instead of the more common sets of relational records. This data structure has been successfully applied in data mining applications ranging from Classification and Clustering with K-Nearest Neighbor, to Classification with Decision Tree Induction, to Association Rule Mining [12][7][19][1][22][2][13][18] [24][6]. A basic P-tree represents one attribute bit that is reorganized into a tree structure by recursive sub-division, while recording the predicate truth value for each division. Each level of the tree contains truth-bits that represent sub-trees and can then be used for phenomena masking and fast computation of counts. This construction is continued recursively down each tree path until downward closure is reached. E.g., if the predicate is “purely 1 bits”, downward closure is reached when purity is reached (either purely 1 bits or purely 0 bits). In this case, a tree branch is terminated when a sub-division is reached that is entirely pure (which may or may not be at the leaf level). These basic P-trees and their complements are combined using boolean algebra operators such as AND(&) OR(|) and NOT(`) to produce mask P-trees for individual values, individual tuples, value intervals, tuple rectangles, or any other attribute pattern[3]. The root count of any P-tree will indicate the occurrence count of that pattern. The P-tree data structure provides a structure for counting patterns in an efficient, highly scalable manner. 2. The Case: Most Classifiers are Near Neighbor Classifiers 2.1 Classification Given a (large) TRAINING SET T(A1, ..., An, C) with CLASS, C, and FEATURES A=(A1,...,An), C Classification of an unclassified sample, (a1,...,an) is just: SELECT Max (Count (T.Ci)) FROM T WHERE T.A1 = a1 AND T.A2 = a2 ... AND T.An = an GROUP BY T.C; i.e., just a SELECTION, since C-Classification is assigning to (a1..an) the most frequent C-value in RA=(a1..an). But, if the EQUALITY SELECTION is empty, then we need a FUZZY QUERY to find NEAR NEIGHBORs (NNs) instead of exact matches. That's Nearest Neighbor Classification (NNC). E.g., Medical Expert System (Ask a Nurse): Symptoms plus past diagnoses are collected into a table called CASES. For each undiagnosed new_symptoms, CASES is searched for matches: SELECT DIAGNOSIS FROM CASES WHERE CASES.SYMPTOMS = new_symptoms; If there is a predominant DIAGNOSIS, then report it, ElseIf there's no predominant DIAGNOSIS, then Classify instead of Query, i.e., find the fuzzy matches (near neighbors) SELECT DIAGNOSIS FROM CASES WHERE CASES.SYMPTOMS ≅ new_symptoms Else call your doctor in the morning That's exactly (Nearest Neighbor) Classification!! CAR TALK radio show example: Click and Clack the Tappet brothers have a vast TRAINING SET on car problems and solutions built from experience. They search that TRAINING SET for close matches to predict solutions based on previous successful cases. (Nearest Neighbor) Classification!! That's We all perform Nearest Neighbor Classification every day of our lives. E.g., We learn when to apply specific programming/debugging techniques so that we can apply them to similar situations thereafter. COMPUTERIZED NNC = MACHINE LEARNING (most clustering (which is just partitioning) is done as a simplifying prelude to classification). Again, given a TRAINING SET, R(A1,..,An,C), with C=CLASSES and (A1..An)=FEATURES Nearest Neighbor Classification (NNC) = selecting a set of R-tuples with similar features (to the unclassified sample) and then letting the corresponding class values vote. Nearest Neighbor Classification won't work very well if the vote is inconclusive (close to a tie) or if similar (near) is not well defined, then we build a MODEL of TRAINING SET (at, possibly, great 1-time expense?) When a MODEL is built first the technique is called Eager classification, whereas model-less methods like Nearest Neighbor are called Lazy or Sample-based. Eager Classifiers models can be: decision trees, probabilistic models (Bayesian Classifier, Neural Networks, Support Vector Machines, etc. How do you decide when an EAGER model is good enough to use? How do you decide if a Nearest Neighbor Classifier is working well enough? We have a TEST PHASE. Typically, we set aside some training tuples as a Test Set (then, of course, those Test tuples cannot be used in model building or and cannot be used as nearest neighbors). If the classifier passes the the test (a high enough % of Test tuples are correctly classified by the classifier) it is accepted. EXAMPLE: Computer Ownership TRAINING SET for predicting who owns a computer: Customer (Age | | | | | 24 58 48 58 28 Salary | | | | | 55,000 94,000 14,000 19,000 18,000 Job | | | | | Owns Computer Programmer | yes Doctor | no Laborer | no Domestic | no Builder | no A Decision Tree classifier might be built from this TRAINING SET as follows: Age < 30 / \ T / Salary>50K / \ T F / Yes 24|55000|Prog|yes F \ No 58|94000|Doctor |no 48|14000|laborer |no 58|19000|Dommestic|no \ \ \ \ \ \No 28|18000|Builder |no The question is: how then is this really a Near Neighbor Classifier (where are Near Neighborhoods involved?)? In actuality, what we are doing is saying that the training subset at the bottom of each decision path represents a near neighborhood of any unclassified sample that traverses the decision tree to that leaf. The concept of “near” or “correlation” used is that the unclassified sample meets the same set of condition criteria as the near neighbors at the bottom of that path of condition criteria. Thus, in a real sense, we are using a different (accumulative) “correlation” definition along each branch of the decision tree and the subsets at the leaf of each branch are true Near Neighbor sets for the respective correlations or notions of nearness. Similarly, for any Neural Network classifier [8][9][16], we train the NN by adjusting the weights and biases through back-propagation until we reach an acceptable level of performance. In so doing we are using the matrix of weights and biases as the determiners of our near neighbor sets. We don’t stop training until those near neighbor sets (the sets of inputs that produce the same class output), are sufficiently “near” to each other to give us a level of accuracy that is sufficient for our needs. 2.2 Boundary based Classification By contrast, it is only fair to say that viewing Support Vector Machine classification as a form of Nearest Neighbor Classification is a stretch. On the other hand, the very first step in SVM classification is often to isolate a neighborhood in which to examine the boundary and the margins of the boundary between classes (assuming a binary classification problem). We will not go into any more detail on the issue of SVM as a NNC method due to size limitations of this paper. 3. CONCLUSIONS AND FUTURE WORK In this paper, we have made the case that classification and prediction algorithms (at least Decision Induction type classifiers and Neural Network type classifiers) are nearest neighbor vote classification and predictions. The conclusion depends upon how one defines “near” and we have shown that there are clearly “nearnesses” or “correlations” that provide these definitions. Two samples are considered near if their correlation is high enough. Broadly speaking, this is the way we always proceed in Classification. This is important because the first decision that needs to be made when faced with a classification or prediction problem is to decide which classification or prediction algorithm to employ. We show how, e.g., Decision Tree Induction, as a classification method can be viewed legitimately as a particular type of Nearest Neighbor Classification. We also will show how Neural Network Predictors are really Nearest Neighbor Classifiers. What good does this understanding do for someone faced with a classification or prediction problem? In a real sense the point of this paper is to head off the standard way of approaching Classification, which seem to be that of using a model-based classification method unless it just doesn’t work well enough and only then using Nearest Neighbor Classification. Our point is that “It is all Nearest Neighbor Classification” essentially and that standard NNC should be used UNLESS it takes too long. Only then should one consider giving up accuracy (of your near neighbor set) for speed by using a model (Decision Tree or Neural Network). 4. REFERENCES [1] Abidin T., and Perrizo W., SMART-TV: A Fast and Scalable Nearest Neighbor Based Classifier for Data Mining. Proceedings of the 21st ACM Symposium on Applied Computing , Dijon, France, April 2006. [2] Abidin, T., Perera, A., Serazi,M., Perrizo,W., Vertical Set Square Distance: A Fast and Scalable Technique to Compute Total Variation in Large Datasets, CATA-2005 New Orleans, 2005. [3] Q. Ding, M. Khan, A. Roy, and W. Perrizo, The P-tree Algebra, Proceedings of the ACM Sym. on App. Comp., pp. 426-431, 2002. [4] Bandyopadhyay, S., and Muthy, C.A., Classification Using Genetic Algorithms. Recognition Letters, Vol. 16, (1995) 801-808. Pattern Pattern [5] Cost, S. and Salzberg, S., A weighted nearest neighbor algorithm for learning with symbolicfeatures, Machine Learning, 10, 57-78, 1993. [6] DataSURG, P-tree Application Programming Interface Documentation, North Dakota State University. http://midas.cs.ndsu.nodak.edu/~datasurg/ptree/ [7] Ding, Q., Ding, Q., Perrizo, W., “ARM on RSI Using Ptrees,” Pacific-Asia KDD Conf., pp. 66-79, Taipei, May2002. [8] Duch W, Grudzi ´N.K., and Diercksen G., Neural Minimal Distance Methods., World Congress of Computational intelligence, May 1998, Anchorage, Alaska, IJCNN’98 Proceedings, pp. 1299-1304. [23] Raymer, M.L. Punch, W.F., Goodman, E.D., Kuhn, L.A., and Jain, A.K.: Dimensionality Reduction Using Genetic Algorithms. IEEE Transactions on Evolutionary Computation, Vol. 4, (2000) 164-171 [9] Goldberg, D.E., Genetic Algorithms in Search Optimization, and Machine Learning, Addison Wesley, 1989. [24] Serazi, M. Perera, A., Ding, Q., Malakhov, V., Rahal, I., Pan, F., Ren, D., Wu, W., and Perrizo,W., DataMIME, ACM SIGMOD, Paris, France, June 2004. [10] Guerra-Salcedo C., and Whitley D., Feature Selection mechanisms for ensemble creation: a genetic search perspective, Data Mining with Evolutionary Algorithms: Research Directions – Papers from the AAAI Workshop, 13-17. Technical Report WS-99-06. AAAI Press (1999). [25] Vafaie, H. and De Jong, K.: Robust feature Selection algorithms. Proceeding of IEEE International Conference on Tools with AI, Boston, Mass., USA. November. (1993) 356-363. [11] Jain, A. K.; Zongker, D. Feature Selection: Evaluation, Application, and Small Sample Performance. IEEE Transaction on Pattern Analysis and Machine Intelligence, Vol. 19, No. 2, February (1997) [12] Khan M., Ding Q., Perrizo W., k-Nearest Neighbor Classification on Spatial Data Streams Using P-trees, Advances in KDD, Springer Lecture Notes in Artificial Intelligence, LNAI 2336, 2002, pp 517-528. [13] Khan,M., Ding, Q., and Perrizo,W., K-Nearest Neighbor Classification of Spatial Data Streams using P-trees, Proceedings of the PAKDD, pp. 517-528, 2002. [14] Krishnaiah, P.R., and Kanal L.N., Handbook of statistics 2: classification, pattern recognition and reduction of dimensionality. North Holland, Amsterdam 1982. [15] Kuncheva, L.I., and Jain, L.C.: Designing Classifier Fusion Systems by Genetic Algorithms. IEEE Transaction on Evolutionary Computation, Vol. 33 (2000) 351-373. [16] Lane, T., ACM Knowledge Discovery and Data Mining Cup 2006, http://www.kdd2006.com/kddcup.html [17] Martin-Bautista M.J., and Vila M.A.: A survey of genetic feature selection in mining issues. ProceedingCongress on Evolutionary Computation (CEC-99), Washington D.C., July (1999) 1314-1321. [18] Perera, A., Abidin T., Serazi, M. Hamer, G., Perrizo, W., Vertical Set Square Distance Based Clustering without Prior Knowledge of K, 14th International Conference on Intelligent and Adaptive Systems and Software Engineering (IASSE'05), Toronto, Canada, 2004. [19] Perera, A., Denton A., Kotala P., Jockheck W., Valdivia W., Perrizo W., P-tree Classification of Yeast Gene Deletion Data, SIGKDD Explorations, Volume 4, Issue 2, December 2002. [20] Perera A. and Perrizo W., Vertical K-Median Clustering, In Proceeding of the 21st International Conference on Computers and Their Applications (CATA-06), March 2325, 2006 Seattle, Washington, USA. [21] Punch, W.F. Goodman, E.D., Pei, M., Chia-Shun, L., Hovland, P., and Enbody, R., Further research on feature selection and classification using genetic algorithms, Proc. of the Fifth Int. Conf. on Genetic Algorithms, pp 557-564, San Mateo, CA, 1993. [22] Rahal, I. and Perrizo, W., An Optimized Approach for KNN Text Categorization using P-Trees. Proceedings. of ACM Symposium on Applied Computing, pp. 613-617, 2004.