The Coding Sequence
advertisement

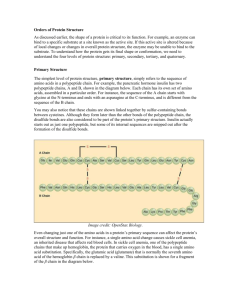
Bioinformatics at the DNALC! In April 2003 it was announced that the final draft sequence of the human genome was complete. This monumental achievement is fueling tremendous research efforts to understand the information our DNA sequence encodes. Scientists have begun to identify genes, define the proteins these genes may produce, and understand how these proteins function. To achieve these goals, biologists are integrating computer-based tools into their research routines. This new field, called bioinformatics, allows scientists to make sense of huge amounts of sequence data and to "mine" genomes for meaning. Students visiting the DNALC will have the unprecedented opportunity to work with the same computer tools and data that genome scientists use. The six computer-based modules listed below integrate enticing content with hands-on computer exercises. Students will analyze human, plant, bacterial, and viral genomes; compare DNA sequences across species; study the evolution of modern humans; understand how variations in DNA sequence contribute to disease; view three-dimensional structures of proteins; and learn about new strategies for developing therapeutic drugs. All classes are two and a half hours in length, and will be conducted in our state-of-the-art Biomedia computer lab. Sickle Cell Anemia – A Disease of Diverse Populations This computer-based lab will explore the molecular biology of sickle cell anemia from DNA sequence, to protein structure, and ultimately to disorder. Students will compare sickle cell gene sequences from patients around the world to elucidate the multiple origins of the disease. Computer simulations will address questions about why the sickle cell mutation continues to persist in several areas of the world. Students will also learn about current and emerging therapies to improve the lives of individuals with this disorder. RESERVATION DETAILS Bioinformatics labs are restricted to students in grades 10, 11, and 12. Each Curriculum Study school is limited to four reservations – one free! – during academic year 2003-04. Non-Curriculum Study schools are limited to three reservations. The group lab rate is $13 per student with a minimum fee of $260. Unless other arrangements have been made in advance, all Bioinformatics labs begin promptly at 9:30 AM. Classes cancelled less than one month prior to their scheduled date will not be permitted additional computer lab visits. Reserve by phone; contact Amanda McBrien at (516) 367-5175. Sickle-Cell Anemia – A Beneficial Genetic Disorder? A Disease Persists – Does Looking at the Distribution Help Understand? Map: distribution of sickle-cell anemia Maps: distribution of malaria and anopheles spec. How DNA Encodes Life DNA RNA Protein Schematic representations in Genome Genome Mining Gene Features Animations on transcription and translation in Code What Is a Genetic Disorder? Example: Sickle Cell Anemia Caused by faulty protein, due to mutated gene What’s a mutation SS is lethal, SA benign. Why does it persist? Map: coincidence of malaria and HBS because HBS provides some resistance for people infected with malaria. How can it be detected? Electrophoresis of proteins: HBS runs faster than HBB AA: 1 large band; AS: 2 bands, 1 large and 1 small; SS: 1 small band Proteins – How/Why They Work Example: Hemoglobin Goal: To find the hemoglobin beta in protein in databases, to examine its structure, and to learn how to handle a protein structure viewer. Physiology: cells need glucose and oxygen to make energy – how do they get oxygen? Red blood cells: circulating; loading oxygen in lungs; unloading oxygen in tissue Why red? Iron. Hemoglobin molecule: consists of four pairwise identical subunits: hemoglobin alpha (HBA) and hemoglobin beta (HBB). Each carries a ring-shaped molecule (porphyrin-ring) which harbors an iron atom to form heme. The iron is the atom that captures the oxygen, the porphyrine anchors iron in the protein, the protein directs the iron to bind and to release the oxygen. What are proteins? Chains of amino acids. How do they work? Because they have a very specific 3D-structure AND because they have very specific amino acids in very specific places. What’s with the amino acids? Let’s look at four: Glu Pro Val A big one!!! The characteristics of the amino acids in proteins direct proteins’ folding AND reaction capacity. Hemoglobin: Usually, iron would just be by itself. However, correctly folded hemoglobinchains are able to harbor porphyrin-rings which, in turn, are capable of holding iron atoms (one per chain). Usually, iron binds oxygen very strongly. However, correctly placed amino acids in the surrounding hemoglobin-chain regulate the ability of iron to hold on to oxygen. Let’s look at cool images of proteins. Go to NCBI (http://www.ncbi.nlm.nih.gov/) Find the words Search Entrez for Change Entrez to Nucleotide Into the search window type HBB homo sapiens mRNA; hit Go. Click on NM_000518 Click on Links (right side of the page) Click on Protein Click on NP_000509 How many amino acids does the protein have? 147 aa Click on BLink Click on 3D structures Find the listing for accession 1KOYB; click on the blue circle for this entry. Click Open Maximize the graphic window (Cn3D 4.1); move the Sequence/Alignment Viewer window to the lower edge of the screen; close the Cn3D Message Log window. HOW TO ZOOM IN AND OUT Left- click on the black background and press z to zoom in. Click on the black background and press x to zoom out one step. Hit x again. HOW TO ROTATE THE MOLECULE To rotate the molecule grab a corner of the molecule with your left mouse button, move your mouse. HOW TO MOVE THE MOLECULE To move the molecule hold Shift down, grab the molecule with your left mouse button, move your mouse right or left, up or down. What different things can you identify in this view? Protein, porphyrin rings, iron, some organic and anorganic molecules. HOW TO CHANGE WHAT’s HIGHLIGHTED Go to Style Click Rendering Shortcuts. Click Worms. Go to Style. Click Coloring Shortcuts. Click …. What different types of structures can you identify in this view? Beta sheets and alpha helices. What’s the protein structure? Hemoglobin beta chain What protein is HBB a part of? hemoglobin How many different iron-bearing structures (porphyrin rings) can you identify? 4 What are they and what function do they serve? Heme-groups; oxygen-transport Four subunits, two alpha and two beta chains. HIDE/SHOW Go to Show/Hide, click on everything that’s not highlighted, click Apply, click Done. What happened? The entire hemoglobin molecule became visible. CHANGE RENDERING Go to Style, Coloring Shortcuts, click Molecule What happened? The four different subunits are shown. Go to Style, Rendering Shortcuts, click Worms. What happened? The two different protein substructures alpha-helices and beta sheets became visible. How did they get to these images? They are determined by x-ray crystallography. Purify and concentrate the protein. Crystallize the protein. Project x-rays through protein and onto film. Develop film. Analyze the patterns of shades and white spaces on the film. Put the puzzle together to see where the atoms are that produced the shades – Voila! You see a protein. Not guess-work but thorough analysis!!! A Mapquest For The Human Genome Goal: To find out where in the human genome hemoglobin genes are located and what the characteristics of the hemoglobin beta gene (HBB) are. Is HBB an ORF gene or a spliced gene? Go to NCBI (http://www.ncbi.nlm.nih.gov/) Find the words Map Viewer; click on them. The genomes of how many different organisms can be accessed through Map Viewer? 19 organisms Find Homo sapiens (human); click on it. How many chromosomes does the human genome consist of? 23, 24, or 25 chromosomes. Justify your response. How many chromosomes are on display? 25 chromosomes Why this number? The human genome consists of 25 separate DNA entities. Find Search for; type hemoglobin into the search window; click Find On what chromosome(s) can you locate entries containing the word hemoglobin? 6, 7, 8, 11, 16, and X Check the list underneath the image and find out on which chromosome hemoglobin beta, HBB is located. Chromosome 11. Click on the link HBB. Use the ruler next to the gene and determine the length of the gene. (Hint: the numbers on the ruler mark nucleotide positions on the chromosome. Use your computers calculator to determine the length of the HBB gene.) 1,600 bp What three different structures can you identify in the cartoon representing the HBB gene? Filled blue box, empty blue box, blue line What do you think the blue boxes represent? Coding sequence that’s translated into protein What the blue lines? introns The Coding Sequence Goal: To isolate and examine the DNA sequence encoding the hemoglobin beta protein. Go to NCBI (http://www.ncbi.nlm.nih.gov/) Find the words Search Entrez for Following the word for type the words hemoglobin homo sapiens Click the button Go How many nucleotide entries did Entrez locate? 9,686 How many protein entries did Entrez locate? 535 How many protein structures did Entrez locate? 107 Find and click Nucleotide: sequence database (GenBank) Does the listing only show entries for humans? If no, which else? No, rat, worm, mustard weed, a bacterium On the page listing the hits find Homo sapiens hemoglobin, beta (HBB), mRNA Click on NM_000518 Study the entry How many basepairs (bp) long is the nucleotide sequence displayed? 626bp At what nucleotide position is the start codon located? That is the position where the coding sequence of the mRNA (CDS) begins. 51 Where does the coding sequence end? 494 How many nucletoides long is the coding sequence? (The result must be a multiple of 3!) 444 Which of the three possible different stop codons TAA, TAG, TGA terminates the CDS? TAA How many aminoacids (aa) is the protein long? 147 What kind of nucleotide polymer is the sequence represent? mRNA How do you explain that the sequence does not contain U’s? To simplify the work with nucleotide sequences databases only use A, C, G, and T, even though RNA does not contain T’s but U’s. This is ok, however, because all you need to know whether a molecule is DNA or RNA. If it’s RNA you could just replace all T’s with U’s to get a realistic representation of the RNA molecule. In order to work with the sequence, transfer it to a database called Sequence Server. Highlight and copy the entire nucleotide sequence from nucleotide 1 through 626. Go to the DNALC BioServers at http://www.bioservers.org/bioserver/ Under SequenceServer click Enter. Close the pop-up Manual. Click CREATE SEQUENCE. Paste the sequence into the Sequence window. Give the sequence some name (e.g. HBB mRNA) and write it into the Name window. Click OK. This is the RNA sequence, it’s 626 bp long. The Gene Goal: To find the HBB gene in human genomic DNA. Identify the gene in the gene sequence (link) by aligning it with the mRNA sequence. Highlight and copy the HBB mRNA sequence from SequenceServer. Open http://pbil.univ-lyon1.fr/sim4.php Paste the sequence into the first window cDNA Sequence. Copy and highlight the genomic DNA sequence (link) and paste it into the lower window Genomic Sequence. Click Submit. Visualize the alignment with LalnView. Which represents the genomic sequence, Seq1 or Seq2? Which the RNA (cDNA)? Seq1 is cDNA, Seq2 is gDNA Which are the introns and exons? Exons are black boxes, introns are empty boxes At which position in the RNA did the coding sequence (CDS) begin? Where was the start codon? Nucleotide 51 Would that be at the beginning of a black box or somewhere within? 51 nucleotides into it So, do exons beging with start codons or do the begin before an actual start codon? Exons begin before the start codon, a start codon is usually located within an exon. The Mutation Goal: To identify the DNA mutation and amino acid change leading to the HBS protein Mutations in DNA: Align the mRNA/coding sequence for HBB and HBS to find out where they differ. Go to http://www.bioservers.org/bioserver/ Under SequenceServer click Enter. Click Manage Groups. In the upper right-hand corner find Sequence Sources, change Classes to Public. Find HBB and Sickle Cell Anemia. Check the box at the LEFT margin, click OK at the bottom of the page. Go to the word None, click on the downward arrow at the right to open pull-down menu. Click HBS CDS, homo sapiens. Go to COMPARE and set the box to the right of it to Align: CLUSTAL W (it may be set already) Click COMPARE to align the two sequences – WAIT! Maximize the result window. How many differences can you find between the two sequences? 4 What are the differences between the two sequences? T/C, A/T, T/A, T/A Which nucleotides are different? (Hint: count the A in the start codon ATG as 1) 9, 20, 172, 387 Which amino acid positions in the protein would these differences effect? 3, 7, 58, 129 Mutations in proteins: Get the amino acid sequences for betaglobin and sickle-cell globin by translating the DNAs. Find HBB cDNA, homo sapiens, click Open. Move the cursor just before the A of the ATG on the third line. Hit Return/Enter on your keyboard, this moves the ATG to the fourth line.. Highlight and copy the sequence from the ATG to the end (don’t worry about the stop codon …). Click Done, In a new browser window open Gene Boy (http://www.dnai.org/geneboy/) Click Your Sequence. Paste the sequence into the workspace. Click Save Sequence (your sequence should have 576 nucleotides). On the Operations panel to the right click Transform Sequence, select Amino Acids. Highlight the sequence under Reading Frame RF1 and copy it. Open the Word program and paste the amino acid sequence into it. Place a carriage return at the end of the sequence. Place a “>” sign in front of the sequence, followed by the letters “HBB”. Type a carriage return. Repeat this process for the sickle cell mRNA (HBS CDS, homo sapiens) with the following modifications: use HBS CDS, homo sapiens instead of HBB cDNA, homo sapiens; copy the entire sequence; pasting this sequence into GeneBoy should yield you 444 amino acids; write HBS before the sequence instead of HBB. Now you should have both amino acid sequences in the Word-file, both preceded by a line that starts out with a “>” followed by the sequence name. Save the file as “betaglobin mutation” Look at the two sequence – why do you think the one for HBB is longer than the one for HBS? Because the nucleotide sequence used t o generate the HBS protein contained the coding sequence exclusively – from start codon to stop codon. The HBB nucleotide sequence contained 132 nucleotides beyond the stop codon. Stops are identified with a star “*”. Examine where the stops within the HBB amino acid sequence are located to identify the end of the HBB protein. It should end with the same amino acids as the HBS protein. Align the two amino acid sequences to identify how they differ: Highlight and copy the content of the Word-file. Go to http://www.ebi.ac.uk/clustalw/ Find Enter or Paste a set of Sequences … and paste the sequence into the box. Click Run and wait until the result is displayed. The result window shows an alignment of the two amino acid sequences. Underneath the alignment is a string of stars denoting identical amino acids. Find the amino acid differences between HBB and HBS. Ignore, however, the end where only HBB shows amino acids; this region is not part of the HBB protein. The HBB as well as the HBS proteins end with the amino acid sequence AHKYH. How many differences can you find between the two amino acid sequences? 1 Counting Met (M) as 1 what are the positions of the differences? Amino Acid 7 What are the differences? Glutamate in HBB vs. valine in HBS How many differences did you find on the DNA level? 4 Where did you expect these differences to be located in the protein? 3, 7, 58, 129 Did any of your expectations turn out right? Yes, position 7 is different. How about the others? DNA variation did not lead to aa variation due to redundancy of genetic code. Effect of mutation on protein structure The mutation obviously has a significant effect on the function of the protein. Let’s see how the exchange from E in HBB to V in HBS affects the protein structure. 1) What is the difference between glutamate (E) and valine (V)? Go to http://info.bio.cmu.edu/Courses/BiochemMols/AAViewer/AAVFrameset.htm Set left window to valine What is the chemical formula for the Valine sidegroup? -CH-(CH3)2 CH3 is not charged, it is not ionic. It’s electrochemically neutral. Is the sidegroup for Valine charged (polar) or not (non-polar)? No, its non-polar, not charged hydrophobic; rejects water Turn the molecule so that the aminoacid-core molecules, the red/blue “V” is positioned on top. Set the right window to glutamate; position red/blue “V” on top. What is the chemical formula for the Glutamate sidegroup? –CH2-CH2-COOH COOH releases an electron to become COO-; it is a charged molecule. So, is the sidegroup for Glutamate charged (polar) or not (non-polar)? Its polar, charged hydrophilic; accepts water Open a new browser window and go back to the protein view Let’s see how the differences between glutamate and valine affect the protein structure The Sickle-Cell Protein Goal: To determine the differences in the structures of hemoglobin beta (HBB) and sickle-cell hemoglobin (HBS) Open this file (link) to view the HBB structure and this file (link) for the HBS structure. Arrange the two images side by side; align the Sequence windows for each structure underneath the respective image screen and adjust the sizes so that they fit side-by-side as well. Close the Message Log windows. Identify in the sequence the amino acid which is different between the two proteins. In both sequences click on this amino acid - this will highlight the amino acid within the protein, too. Moving the proteins about, can you identify any differences in the protein structures? Zoom into the image and center on the highlighted amino acid. Under Style Rendering Shortcuts choose Toggle Sidechains – can you identify the differences in the sidegroup structures for Valine (V) in HBS and Glutamate (E) in HBB? View an alignment of the HBB and HBS Proteins Go to NCBI (http://www.ncbi.nlm.nih.gov/) Find the words Search Entrez for Change Entrez to Structure Into the search window type HBS; hit Go. Click on 2HBS Click on the term Chain B (find the blue bar …) Click on View 3D Structure Click on Open. Maximize the Cn3D screen; align the Sequence screen underneath; close the Message/Log screen. How different are the two proteins? Not at all. Identify and highlight in both sequences the amino acid that’s different. Can you see a difference now? Nope Go to Style, Rendering Shortcuts, click Toggle Sidechains. Make sure the V and E in position 6 of both sequences are highlighted. Can you see a difference now? Sure Change the highlighting from position 6 to position 5 (Proline; P). Can you see a difference now? Nope. The Effect of the Mutation The change from glutamate to valine in position 7 of the hemoglobin beta chain does not affect the capability of the molecule to transport oxygen. The introduction of valine in place of glutamate introduces a hydrophobic molecule into the position that was previously occupied by a hydrophilic molecule. This generates a site that tries to minimize contact with water (or hydrophilic molecules) – by means of connecting to other hydrophobic amino acids. If the protein is loaded with oxygen the hemoglobin protein is twisted in a fashion that turns the valine to the inside of the protein, covering it from the surrounding hydrated environment – everything is as in the regular hemoglobine molecule. Release of oxygen, however, leads to a change in the 3D-structure exposing the valine to the surrounding hydrated environment. This generates a sort of “sticky-ness”, a region which is available to connect with other hydrophobic (or at least neutral) amino acids in order to form a local niche from which water is excluded. And Jennie is going to show you how that causes sicklecell hemoglobin to malfunction and cause discomfort and disease. The Disease RESOURCES WWW Resources: The Mistery of the Crooked Cell at the University of North Carolin at http://www.unc.edu/cell/files/extensions/mystery/mystery.html The Sickle Cell Information Center at http://www.scinfo.org/toc.htm Sickle Cell Disease Association of America (SCDAA) at http://www.sicklecelldisease.org/ The American Sickle Cell Disease Association (ASCDA) at http://www.ascaa.org/ National Heart, Lung, and Blood Institute at http://123819272.net/Diseases/Sca/SCA_WhatIs.html NIH’s Medline at http://www.nlm.nih.gov/medlineplus/sicklecellanemia.html NCBI’s Online Mendelian Inhertiance in Man (OMIM) at http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=603903 NCBI’s Genes and Disease resources at http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View..ShowSection&rid=gnd.section.98 The Humane Genome Sequencing Project at http://www.ornl.gov/TechResources/Human_Genome/posters/chromosome/sca.html Howard University’s Center for Sickle Cell Disease at http://www.huhosp.org/sicklecell/ Harvard University’s Information Center for Sickle Cell and Thalassemia Disorders at http://sickle.bwh.harvard.edu/ WHO’s Malaria Information at http://www.who.int/tdr/diseases/malaria/diseaseinfo.htm CDC’s Malaria Information at http://www.cdc.gov/travel/malinfo.htm The Museum of South Africa’s Information on Malaria, Anopheles, and Plasmodium at http://www.museums.org.za/bio/apicomplexa/plasmodium.htm