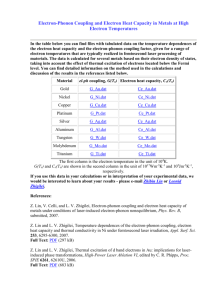
Local Tomography Software, LOTOS-06 - ivan
advertisement

Active source tomography algorithm
for 3D inversion, ATOM-3D
(version of November 2008)
Contact person: Ivan Koulakov (KoulakovIY@ipgg.nsc.ru)
Content
1. General structure of ATOM-3D
1.1. List of folders in the root directory
1.2. Structure of the DATA folder
1.2.1. Organization of the input data in the "inidata" folder.
1.2.2. Organization of the MODEL folder.
1.2.3. Description of some of the main initial parameters.
2. Description of the ATOM-3D algorithm
2.1. Iterative tomographic inversion.
2.2. Ray tracing in the 3D velocity model
2.3. Construction of the parameterization grid (nodes)
2.4. Calculation of matrix
2.5. Inversion
2.6. Calculation of 3D model in a regular grid
2.7. Practical realization of LOTOS-07 code for the real data inversion
3. Presentation of the results
3.1. Horizontal sections of the resulting anomalies
3.2. Vertical sections of the resulting anomalies.
4. Synthetic modeling
4.1. General remarks
4.2. Definition of noise
4.3. Visualization of the initial synthetic models
4.4. Definition of checkerboard anomalies
4.5. Definition of free horizontal anomalies
4.6. Definition of free vertical anomalies
4.7. Definition of vertical checkerboard anomalies
4.8. Practical realization of LOTOS-07 code for synthetic tests
A tomographic algorithm, ATOM-3D (Active TOMography in 3D) is designed for
investigating 3D velocity structure based on travel times of first arrival refracted seismic
rays from active sources. Both marine and land observation schemes can be considered.
The ATOM-3D code can be directly applied to very different data sets without
complicated tuning of parameters. It has a quite wide range of possibilities for performing
different test and is quite easy to operate. ATOM-3D code is freely available online at
www.ivan-art.com/science/ATOM_3D. Any help with installation and running the code
can be obtained from the author, Ivan Koulakov (KoulakovIY@ipgg.nsc.ru).
1. General file structure of ATOM-3D
1.1. List of folders in the root directory
The root forlder, ATOM_3D/ contains the following folders (some of them are
obligatory and some are optional):
1. DATA (fixed format): folder which contains all the data and models
2. FIG_files (fixed format): folder for files generated at the step of visualization
which can be then viewed in Surfer or similar visualization programs.
3. PICTURE (free format, optional): folder for storing the resulting images.
4. PROG (fixed format): folder with all programs for iterative, nonlinear
tomographic inversion.
5. subr (free format): folder which contains all the subroutines.
6. tmp (fixed format): folder for temporary files. Initially it can be empty.
7. VISUAL (free format): folder for visualization programs.
8. Files in the root folder are used for prompt definition of the inversion procedure.
1.2. Structure of the DATA folder
Table 1. Hierarchy of the DATA folder structures in ATOM-3D
root data folder
area folder
model folder
inidata
MODEL_01
DATASET1
BOARD_01
LOTOS_01
DATA
inidata
MODEL_01
DATASET2
MODEL_02
SYN_MOD1
Hierarchy of the DATA folder structure is shown in Table 1. The DATA contains the
Area folders (e. g. “DATASET1”). The name of the Area folder consists of 8 characters.
Each “Area” folder contains “inidata” folder with initial data and folders with real
data or synthetic models.
Also it may contain any number of MODEL folders with real data and synthetic
models. The name of each model should consist of 8 characters.
1.2.1.Organization of the input data in the "inidata" folder.
The input data are contained in the “inidata” folder and include two files: rays.dat
and topo.grd
1. “rays.dat”: list of all source-receiver pairs and travel times
Each line of “rays.dat” contains 7 numbers with free format which correspond to one
ray:
longitude (degree, W-negative), latitude (degrees, S-negative) and depth (km,
positive – below sea level) of sources
longitude (degree, W-negative), latitude (degrees, S-negative) and depth (km,
positive – below sea level) of receivers
observed travel time (in seconds)
It can be read with the following Fortran command:
read(1,*,end=2)fi1,tet1,z1,fi2,tet2,z2,time
Example 1: File “ATOM_3D\DATA\DATASET1\inidata\rays.dat”
-16.22446
0.1997948
-16.23648
0.1997948
-16.24850
0.1997948
-16.26051
0.1997948
-16.27253
0.1997948
-16.28454
0.1997948
-16.29656
0.1997948
-16.30857
0.1997948
-16.32059
0.1997948
-16.33261
0.1997948
-16.34258
0.1997948
-16.35256
0.1997948
28.61077
13.30453
28.60994
13.21514
28.60912
13.07373
28.60830
12.97341
28.60747
12.86394
28.60665
12.74866
28.60583
12.63654
28.60501
12.51454
28.60418
12.40363
28.60336
12.29324
28.59628
12.03732
28.58921
11.75637
0.7753800
-16.84785
28.30860
0.8710325
-16.84785
28.30860
0.9674537
-16.84785
28.30860
1.054933
-16.84785
28.30860
1.137342
-16.84785
28.30860
1.219888
-16.84785
28.30860
1.319931
-16.84785
28.30860
1.420617
-16.84785
28.30860
1.520212
-16.84785
28.30860
1.619736
-16.84785
28.30860
1.522545
-16.84785
28.30860
1.393697
-16.84785
28.30860
2. “topo.grd”: topography-bathymetry in the study area. This file is the standard
grid file used for presenting contours in SURFER software. Actually, this file is
used only for presenting the results in vertical profile and is not necessarily
required. During calculations this file is not used. The format can be described
with following program fragment of a Fortran code:
open(1,file='../../data/'//ar//'/inidata/topo.grd',status='old',err=234)
read(1,*)
read(1,*)nfmap,ntmap
read(1,*)fmap1,fmap2
read(1,*)tmap1,tmap2
read(1,*)zmin,zmax
do itet=1,ntmap
read(1,*)(topo(ifi,itet),ifi=1,nfmap)
end do
close(1)
The topography/bathymetry values are given in meters; negative values are below sea
level.
Example 2: File “ATOM_3D\DATA\DATASET1\inidata\topo.grd”
DSAA
92
-18.03333
27.00000
-3855.000
-3760.000
-3747.000
-3721.000
-3694.000
-3688.000
-3680.000
-3656.000
-3635.000
-3597.000
-3620.000
-3554.000
-3521.000
-3489.000
-3426.000
-3360.000
-3280.000
62
-14.99999
29.03333
3015.000
-3772.000
-3736.000
-3716.000
-3698.000
-3688.000
-3675.000
-3652.000
-3636.000
-3540.000
-3577.000
-3546.000
-3517.000
-3480.000
-3410.000
-3339.000
-3269.000
not important
nfmap, ntmap
fmap1, fmap2
Longitude limits
tet1, tet2
Latitude limits
z1, z2 (not important)
-3771.000
-3741.000
-3730.000
-3723.000
-3707.000
-3700.000
-3695.000
-3688.000
-3686.000
-3684.000
-3669.000
-3664.000
-3649.000
-3645.000
-3660.000
-3700.000
-3566.000
-3609.000
-3563.000
-3561.000
-3536.000
-3530.000
-3509.000
-3500.000
-3467.000
-3453.000
-3393.000
-3382.000
-3334.000
-3317.000
-3245.000
-3213.000
-3733.000
-3723.000
-3697.000
-3687.000
-3682.000
-3661.000
-3639.000
-3674.000
-3627.000
-3560.000
-3527.000
-3494.000
-3439.000
-3378.000
-3293.000
-3193.000
The third file “S_R_pairs.dat” is not strictly required. In this case it was used to generate
the synthetic dataset, which then was used as the main dataset for inversion)
1.2.2.
Organization of the MODEL folder.
The MODEL folder is created either for real data or synthetic tomographic models.
The name of the MODEL folder should contain 8 characters (e.g. “BOLIVAR1”,
“MODEL_01”, “BOARD_01”). The structure of the model folders is shown in Figure
1.1
A real data MODEL folder contain four sub-folders: “grids”, “ini_param”, “result”,
“times”. Initially, only one of them, “ini_param”, is not empty and contains initial
parameters of the model. It is the only one which is required. The other folders can be
empty. If they contain some files (e.g. copied from another model), they will be
overwritten during calculation of new model. If these folders do not exist, the algorithm
creates them automatically
1.2.3.
Description of some of the main initial parameters.
Figure 1.1 Structure of the folder with initial parameters. Examples of “ref_start.dat” and
“MAJOR_PARAM.DAT’ are shown.
Example 3: File “ATOM_3D\DATA\DATASET1\BOLIVAR1\ini_param\MAJOR_PARAM.DAT”
********************************************************
AREA_CENTER :
-16.6 28.3
Center of conversion to XY
********************************************************
Parameters for tracing in 3D model using bending tracing
********************************************************
TRACING_PARAMETERS:
! Parameters for BENDING:
0.5
ds_ini: basic step along the rays
3
min step for bending
0.01
min value of bending
5
max value for bending in 1 step
1
k_reduce: frequency of data to be selected
********************************************************
ORIENTATIONS OF GRIDS :
4
number of grids
0 22 45 67
orientations
********************************************************
INVERSION PARAMETERS :
40
LSQR iterations
0.2
smoothing damping
0.0
amplitude damping
********************************************************
Parameters for 3D model with regular grid
********************************************************
3D_MODEL PARAMETERS:
-40. 40. 0.5 xx1, xx2, dxx,
-40. 40. 0.5 yy1, yy2, dyy,
-5. 20. 0.5 zz1, zz2, dzz
3
distance from nearest node
0
Smoothing factor1
********************************************************
Parameters for grid construction
********************************************************
GRID_PARAMETERS:
-40. 40. 1.0
grid for ray density calculation (X)
-40. 40. 1.0
grid for ray density calculation (Y)
-5. 20. 0.5
min and max levels for grid
1
! Grid type: 1: nodes, 2: blocks
0.5
!min distance between nodes in vert. direction
0.05 100.0
!plotmin, plotmax= maximal ray density, relative to average
-5.
!zupper: Uppermost level for the nodes
0.05
0.05
!dx= step of movement along x
!dz= step of movement along z
The main files with the initial parameters are shown in Figure 1.1 and Example 3. Most
of the parameters for tracing and inversion are defined in ‘MAJOR_PARAM.DAT’. The
content of this file is organized by groups. Each group starts with a key line. For
example:
AREA_CENTER :
ORIENTATIONS OF GRIDS :
INVERSION PARAMETERS :
etc.
After the key line there is a list of parameters for the current group with a fixed format.
The order of groups and number of empty lines between groups are free.
For example, the first group in the presented file is:
********************************************************
AREA_CENTER :
-16.6 28.3
Center of conversion to XY
This parameter group contains the geographical coordinate of the central point of the
study area which is used as a reference point for conversion to Cartesian coordinates.
The other parameters in “MAJOR_PARAM.DAT” will be explained during description
of the main steps.
Starting velocity is 1D distribution which is defined in “refmod.dat”.
Example 4: File “ATOM_3D\DATA\DATASET1\BOLIVAR1\ini_param\refmod.dat”
1.7
0.0000000E+00
4.735601
3.071527
10.00000
20.00000
30.00000
260.0000
6.035635
7.07262
8.063499
8.482500
3.248399
3.445227
3.955563
4.609000
This file contains the information about starting reference velocity model. This format
comes from the LOTOS code where both P and S velocities were investigated.
Although, in ATOM-3D code only one velocity is used, we conserve the same format.
The second column which indicates the S velocity is not used. First line is the Vp/Vs
ratio. If it is zero, the S-velocity is defined according to third column of the following
lines. Otherwise, the S velocity is computed from P velocity (second column) using
constant value of Vp/Vs ratio. Velocities are defined at some depth levels (first column)
and linearly interpolated in between
The other parameters will be presented in description of the ATOM-3D algorithm, next
section.
2. Description of the ATOM-3D algorithm
2.1. Iterative tomographic inversion.
The actual version of LOTO-07 allows performing inversion using two different methods
of parameterization.
Inversion consists of consequent execution of the following projects:
\PROG\1a_resid\
\PROG\2_ray_density\
\PROG\3_grid\
\PROG\4_tetrad\
\PROG\5_sosedi\
\PROG\6_matr\
\PROG\7_invers\
\PROG\8_3D_model\
2.2. Ray tracing in the 3D velocity model
Project: \PROG\1a_resid\
Main INPUT files:
in 1st iteration:
/data/DATASET1/MODEL_01/inidata/rays.dat
in other iterations:
/data/DATASET1/MODEL_01/times/rays_a’//it-1//’.dat
Main OUTPUT file:
/data/DATASET1/MODEL_01/times/rays_a’//it//’.dat
(‘it’ is the number of iteration)
The calculations are controlled by parameters in the file:
File “ATOM_3D\DATA\DATASET1\BOLIVAR1\ini_param\MAJOR_PARAM.DAT”
********************************************************
Parameters for tracing in 3D model using bending tracing
********************************************************
TRACING_PARAMETERS:
! Parameters for BENDING:
0.5
ds_ini: integration step along the rays
3
bend_step: minimal step for bending
0.01
bend_min: minimal value of bending
5
bend_max: maximal value for bending in 1 step
1
k_reduce: frequency of data to be selected
Description of the main principle for the ray tracing
One of the key features of the ATOM-3D code is a ray tracing algorithm based on
the Fermat principle of travel time minimization. A similar approach is used in other
algorithms (e.g., Um and Thurber (1987)) and is called bending tracing. We present our
own modification of the bending algorithm. An important feature of this algorithm is that
it can use any parameterization of the velocity distribution. It is only necessary to define
uniquely one positive velocity values at any point of the study area. It can be done with
nodes or cells, with polygons or analytical laws, or any other ways. The current version
of ATOM-3D includes many various options for velocity definition. However, if
necessary, any other parameterization can be easily included.
A basic principle of our bending algorithm is shown in Figure 1.2. Searching a
path with minimum travel time is performed in several steps. The starting ray path is a
straight line. In the first step (Plot A), the ends of the rays are fixed (points 1 and 2), and
point A in the center of the ray is used for bending. Deformation of the ray path is
performed perpendicular to the ray path in two directions: in and across the plane of the
ray. The values of shift, B, of the new path with respect to the previous one depend
linearly on the distance from A to the ends of the segment, as shown in Figure 1.2. The
value of B is adjusted to obtain the curve γ(B) which provides the minimum value of the
integral:
ds
t
[1]
V
(
s
)
(B)
where V(s) is the velocity distribution along the ray. (ds=dstep). B is varied from a
maximum value, bend_max, to minimum value, bend_min.
In the second step (Plot B), three points are fixed (points 1, 2, and 3), and
deformation of the ray path is performed in two segments (at points A and B). In a third
step (Plot C), four points are fixed and three segments are deformed. In Plot D, the results
of bending are shown for eight segments. The iterations stop when the length of segments
becomes smaller than a predefined value bend_step.
The ray constructed in this way tends to travel through high-velocity anomalies
and avoids low velocity patterns. It should be noted that although a 2D model is shown in
Figure 1.2, the algorithm is designed for the 3D case.
k_reduce is frequency of data which are taken into consideration (e.g. for k_reduce=2
only half of data is used, 3 – only a third part).
Figure 1.2. Grounds of the bending algorithm. Ray construction is demonstrated for a model with
exaggerated velocity contrasts. 1D velocity varies from 2500 to 9000 m/s at 2000 m depth. Hatched light
grey patterns represent negative anomalies of -30%; dark grey patterns are positive anomalies of +30%.
Details of the bending algorithm are given in the text.
Figure 1.3. For the grounds of the sea correction introduction
In the case of marine experiments when sources are located on the sea surface, the travel
time are corrected for the sea column. In this case the full tracing is not performed (in
contrast to the 2D version of the algorithm, PROFIT, where tracing is performed from the
source on the sea surface).
In the case of creating synthetic data (e.g. project “b_synth_times”) tracing is performed
from the point B located on the sea bottom directly beneath source (green line). Then the
travel time of the synthetic ray passing through A-C-E (red line) is computed as:
dZ 1 p 2V02
TACE TBF TBD TAC TBF
[2]
V0
sin 1
where p is the ray parameter: p
, TBF is the computed travel time of the ray BF,
V1
dZ is the depth of the sea, V0 and V1 are velocities in the sea and in just beneath the sea
bottom, respectively.
In the first iteration of the inversion procedure, we replace the source from the point A to
C. Doing this, we assume that 1 is close to 90°. In this case, horizontal shift will be:
BC
dZ V0
[3]
V12 V02
and corrected travel time along the ray CD (blue line) is computed as:
obs
obs
TCE
TACE
dZ V1
[4]
V0 V12 V02
In the next iterations the source is always put in the point C, and the correction is not
applied anymore. The residuals are computed as:
obs
md
dt TCE
TCE
[5]
md
where TCE
is the travel time computed in the current 2D model along the path CE.
2.3. Construction of the parameterization grid (NODES):
Executed Projects:
\PROG\2_ray_density\
\PROG\3_grid\
\PROG\4_tetrad\
\PROG\5_sosedi\
The calculations are controlled by parameters in the file:
Example 3: File “ATOM_3D\DATA\DATASET1\BOLIVAR1\ini_param\MAJOR_PARAM.DAT”
********************************************************
ORIENTATIONS OF GRIDS :
4
number of grids
0 22 45 67
orientations of grids, degrees
********************************************************
Parameters for grid construction
********************************************************
GRID_PARAMETERS:
-40. 40. 1.0
xgr1, xgr2, dxgr: grid for ray density calculation (X)
-40. 40. 1.0
ygr1, ygr2, dygr: grid for ray density calculation (Y)
-5. 20. 0.5
zgr1, zgr2, dzgr: min and max levels for grid
1
! Grid type: 1: nodes (other options are not valid)
0.5
dz_min: min distance between nodes in vert. direction
0.05 100.0
dens_min, dens_max: min and max ray density, relative to average
-5.
z_upper: Uppermost level for the nodes
0.05
0.05
dx_step: step of movement along x
dz_step: step of movement along z
Selected are the most important parameters which determine the vertical and horizontal
spacing of the grid.
The steps of grid construction are following:
1. Ray density calculation: \PROG\2_ray_density\
Summary ray length is computed in cells with spacing dxgr, dygr, dzgr and limits xgr1,
xgr2, ygr1, ygr2, zgr1, zgr2. Then the ray density function is normalized with respect to
the average value in non-empty cells. If the ray density is less than dens_min of the
normalized density, these cells are taken off from consideration. If the ray density is
more than dens_max of the normalized density, the ray density in this cell will be set
equal to dens_max.
2. Installing the nodes: \PROG\3_grid\
The regular nodes with spacing, dxgr, dygr, indicate the vertical lines where the nodes
are installed irregularly according to the ray density. We move along the vertical lines
with the step of dz_step and integrate the ray density function. As soon as it becomes
greater than a predefined value, we put the node, and the integration starts anew. At the
same time, the spacing between the nodes in vertical direction should be not less than
dz_min.
3. Joining the nodes into tetrahedral blocks: \PROG\4_tetrad\
This step is required only for the next step
4. Finding the neighboring nodes: \PROG\5_sosedi\
This step finds all the neighboring nodes in the grid. The pairs of nodes are used in the
smoothing block during inversion
It is important to note that in our algorithm the resolution of the model does not depend
on the grid spacing. It is merely controlled by smoothing and regularization parameters
during the matrix inversion which is described below. However, since the nodes are
placed on planes having a predefined orientation, this can bring some artifacts to the
result of the inversion. To reduce the effect of grid orientation we perform the inversion
in four differently oriented grids (0˚, 22˚, 45˚ and 67˚) and then stack them. Orientations
of grids are defined in “MAJOR_PARAM.DAT”:
********************************************************
ORIENTATIONS OF GRIDS :
4
number of grids
0 22 45 67
orientations of grids, degrees
2.4. Calculation of matrix:
Project \PROG\6_matr\
Matrix calculation is performed along the rays computed by the bending method after the
2.2. The effect of velocity variation at each node on the travel time of each ray (∂t/∂V) is
computed numerically, as in (Koulakov et al., 2006). The data vector corresponding to
this matrix consists of residuals obtained after the step of source location.
2.5. Inversion:
Project \PROG\7_invers\
The parameters for the inversion are contained in file:
File “ATOM_3D\DATA\DATASET1\BOLIVAR1\ini_param\MAJOR_PARAM.DAT”
********************************************************
INVERSION PARAMETERS :
40
N_LSQR: LSQR iterations
0.2
W_smth: smoothing damping
0.0
W_amp: amplitude damping
To control smoothing and amplitude of the 3D velocity models, the matrix obtained in
the step 2.4 is supplemented with special blocks. Each line in the smoothing block
contains two non-zero elements with opposite signs, corresponding to neighboring
parameterization nodes in the model. The data vector corresponding to this block is zero.
Increasing the weight of these elements (W_smth) has a flattening effect upon the
resulting anomalies. The block which controls the amplitude of the model has diagonal
structure with only one element in each line and zero values in data vector (controlled by
W_amp). The resulting matrix is inverted using the LSQR method (Paige and Saunders,
1982; van der Sluis and van der Vorst, 1987). The number of LSQR iterations providing a
satisfactory convergence in our case is N_LSQR.
2.6. Calculation of 3D model in a regular grid:
Project \PROG\7_invers\
After performing the inversions for several grids with different orientations, the velocity
anomalies are recomputed in a 3D regular grid. Parameters of the calculation are defined
in
File “ATOM_3D\DATA\DATASET1\BOLIVAR1\ini_param\MAJOR_PARAM.DAT”
********************************************************
Parameters for 3D model with regular grid
********************************************************
3D_MODEL PARAMETERS:
-40. 40. 0.5
xx1, xx2, dxx: limits and spacing of reg. grid along X
-40. 40. 0.5
yy1, yy2, dyy: limits and spacing of reg. grid along Y
-5. 20. 0.5
zz1, zz2, dzz: limits and spacing of reg. grid along Z
3
d_to_node: distance from nearest node
0
k_smooth: Smoothing factor1
Limits of the volume for interpolation and grid spacing along X, Y and Z are defined in
first three lines. d_to_node means the minimal distance to the nearest parameterization
node of one of the used grids. If the distance is larger, this point is outside the resolved
area and the value there is presumed 0. The algorithm allows smoothing of the velocity
anomalies which is controlled by integer value of k_smooth.
2.7. Practical realization of ATOM-3D code for the real data
inversion
To perform the successful run of the ATOM-3D for the real data, the data structure should
be created, as described in section 1. There is a possibility to run the steps presented in
section 2 manually, step by step. However, the ATOM-3D code contains a program which
performs automatic managing of all steps. The source of this program is presented below:
Program for automatic managing of the ATOM-3D steps:
File “ATOM_3D\PROG\START_real_inversion\start_real.f90”
(the program steps are highlighted with red)
USE DFPORT
character*8 ar,ar_all(10),md,md_all(10),line
character*1 rg_all(100),rg,it
integer kod_loc(10),kod_iter(10),kod_oe(10)
open(1, file='../../all_areas.dat')
do i=1,4
read(1,*)
end do
do i=1,10
read(1,'(a8,1x,a8,1x,i1,1x,i1,1x,i1)',end=7)ar_all(i),md_all(i),kod_iter(i),kod_oe(i)
end do
7 close(1)
n_ar=i-1
! Check all the areas:
do iar=1,n_ar
ar=ar_all(iar)
md=md_all(iar)
niter=kod_iter(iar)
koe=kod_oe(iar)
open(11,file='../../model.dat')
write(11,'(a8)')ar
write(11,'(a8)')md
write(11,'(i1)')1
write(11,'(i1)')1
write(11,'(i1)')koe
close(11)
!******************************************************************
open(1,file='../../data/'//ar//'/'//md//'/ini_param/MAJOR_PARAM.DAT')
do i=1,10000
read(1,'(a8)',end=573)line
if(line.eq.'ORIENTAT') goto 574
end do
573 continue
write(*,*)' cannot find ORIENTATIONS in MAJOR_PARAM.DAT!!!'
pause
574 read(1,*)nornt
close(1)
! Execute the inversion for grids with different orientations:
do iter=1,niter
write(it,'(i1)')iter
open(11,file='../../model.dat')
write(11,'(a8)')ar
write(11,'(a8)')md
write(11,'(i1)')iter
write(11,'(i1)')1
write(11,'(i1)')koe
close(11)
write(*,*)'
Residuals for active data:'
i=system('..\1a_resid\resid.exe')
999
continue
do igr=1,nornt
open(11,file='../../model.dat')
write(11,'(a8)')ar
write(11,'(a8)')md
write(11,'(i1)')iter
write(11,'(i1)')igr
close(11)
if(iter.eq.1) then
write(*,*)'
Compute the ray density'
i=system('..\2_ray_density\plotray.exe')
write(*,*)'
Compute the parameterization grid:'
i=system('..\3_grid\grid.exe')
i=system('..\4_tetrad\Tetrad.exe')
i=system('..\5_sosedi\add_matr.exe')
end if
i=system('..\6_matr\matr.exe')
i=system('..\7_invers\Invbig.exe')
end do
i=system('..\8_3D_model\mod_3D.exe')
end do ! Different grids
end do ! Different areas
stop
end
This program allows running all the ATOM-3D steps for one or several models. The list
of models is defined in file “/ATOM_3D/all_areas.dat”. Example of this file is
presented below:
File “ATOM_3D\all_areas.dat”
1-2: codes of the area and models (each contain 8 characters)
3: number of iterations
4: 0 all data, 1 odd events, 2 even events
**********************************************
DATASET1 MODEL_01 5 0
DATASET1 MODE_ODD 5 1
DATASET1 MODE_EVE 5 2
In the presented example, three models are defined. All of them are from the same AREA
folder, “DATASET1”, indicated in 1st column. First model is “MODEL_01” that is
indicated in 2nd column. It runs for five iterations (indicated in 3rd column). 4th column is
an indicator whether all data are considered or only halved subsets with odd (1) or even
(2) number of events. So, the next two models MODE_ODD and MODE_EVE will be based
on subsets with odd and even numbers of events, respectively. It is important to define all
the parameters in the file “all_areas.dat” according to a fixed format:
(a8,1x,a8,1x,i1,1x,i1) and they should start from line 5. Any number of different models
can be defined. They will run consequently one after another.
3. Presentation of the results
To control realization of intermediary steps and to visualize the final results, some special
program should be run. They produce the files in the “/FIG_file” folder, in corresponding
subfolders, which can be presented in Surfer or other similar visualization software.
3.1. Horizontal sections of the resulting anomalies.
The program which produces the GRD files of the resulting anomalies of P and S
velocities based on node parameterization in horizontal sections is located in the Project:
/VISUAL/hor_result
The general information about the visualized model is presented in file
/VISUAL/area.dat
File “ATOM_3D\VISUAL\area.dat”
DATASET1
MODEL_01
area
model
5
1 4
0
n_iter: iteration
kgr1, kgr2: first and last grids
add_perc: additional bias (in percents) to shift the color scale
Other parameters for visualization are defined in the file “…/ini_param/sethor.dat” (see
example below).
File “ATOM_3D\DATA\DATASET1\MODEL_01\ini_param\sethor.dat”
5
0 3 6 9 12
-0.4 0.4 0.005
3
0
N_lev: number of depth levels
Z_lev(i): Depths of sections
-0.4 0.4 0.005 fi1, fi2, dfi, tet1, tet2, dtet: in degrees
d_to_node: distance from nearest node
k_smooth: Smoothing factor1
In this example, visualization is performed for the model MODEL_01 from the area
“DATASET1” after 5 iteration. The results obtained separately in four different grids,
1,2,3 and 4, will be combined in one map. The results will be presented in five depth
sections at 0, 3, 6, 9 and 12 km. The limits of the map are computed with respect to the
center of the area (-16.6° and 28.3°) defined in AREA_CENTER in
MAJOR_PARAM.DAT. In this case, the map longitudes are between -17º and -16.2º, and
the latitudes are between 27.9º and 28.7º. Steps along longitude and latitude are 0.005º.
The resulting anomalies are only shown only in resolved areas where there are
parameterization nodes. d_to_node means the minimal distance to the nearest
parameterization node of one of the used grids. If the distance is larger, this point is
outside the resolved area and the value there is presumed (dv=-999). The algorithm
allows smoothing of the velocity anomalies which is controlled by integer value of
k_smooth.
The output of this program:
“ATOM_3D/FIG_files/hor/dv_M.grd“: relative anomalies in percent in horizontal
sections. M is a number of depth level according to the information in “sethor.dat“. This
file can be directly visualized in the Surfer Software as a contour line plot.
Resulting velocity anomalies at 0 and 6 km depth (levels 1 and 3) after 5 iteration
corresponding to the model DATASET1/MODEL_01 are shown in Figure 3.1.
Figure 3.1. Resulting velocity anomalies at 0 and 6 km depth (levels 1 and 3) after 5
iteration corresponding to the model DATASET1/MODEL_01. Velocity anomalies are
given in percent with respect to the reference model. Profiles with sources are shown
with black dots (lines).
3.2. Vertical sections of the resulting anomalies.
The program which produces the GRD files of the resulting anomalies and absolute
values of P and S velocities in vertical sections is in a Project: /VISUAL/ver_result
The general information about the visualized model is defined in file
/VISUAL/area.dat
File “ATOM_3D\VISUAL\area.dat”
DATASET1
BRD_VER1
5
1 4
0
area
model
n_iter: iteration
kgr1, kgr2: first and last grids
add_perc: additional bias (in percents) to shift the color scale
Other parameters for visualization are defined in the file “…/ini_param/setver.dat” (see
example below).
File “ATOM_3D\DATA\DATASET1\BRD_VER1\ini_param\setver.dat”
1
N_prof: Number of different sections
-17 28.21 -16.3 28.21 Fi_A,Tet_A,Fi_B,Tet_B: coordinates of ends of the profile A-B
10 distance from section for visualization of events (not used anymore)
0.5
dx_prof: step in horizontal dir
-5 20 0.5
zmin,zmax,dz: depth limits and step along Z
10
ds_mark: Marks for indication of position of section
1
d_to_node: distance from nearest node
0
k_smooth: Smoothing factor1
In this example, visualization is performed for the model MODEL_01 from the area
“BRD_VER1” after 5 iterations. The results obtained separately in four different grids
will be combined in one map. The results will be presented in one cross section. The
coordinates of ends of the sections are defined by fi_A, tet_A, fi_B, tet_B. Horizontal
step along the profiles is defined by dx_prof (0.5 km, in this example). Vertical limits
and step are defined by zmin, zmax and dz.
The resulting anomalies are only shown only in resolved areas where there are
parameterization nodes. If the distance to the nearest node of one of considered grids is
more than d_to_node (1 km, in this example), the value of anomaly is not shown (dv=999).
Visualization program allows smoothing of the visualized map which is controlled by the
k_smooth parameter.
Output of this program:
1. “ATOM_3D/FIG_files/vert/ver_M.grd“: relative anomalies in percent in vertical
sections. M is a number of profile. It can be directly visualized in the Surfer Software as a
contour line plot.
2. “ATOM_3D /FIG_files/vert/abs_M.grd“: absolute velocities in km/s in vertical
sections. M is a number of profile. It can be directly visualized in the Surfer Software as a
contour line plot.
3. “ATOM_3D/FIG_files/vert/mark M.bln“: Position of the profile in the map view.
M is a number of profile. It can be directly visualized in the Surfer Software as a baseline
plot. “mark M.dat” allows visualization of labels along the map view of the profile (e.g.
50, 100, 150...).
Figure 3.2. Resulting velocity anomalies corresponding to “DATASET1/BRD_VER1” after 5 iterations in
map view and a cross section. Velocity anomalies are given in percent with respect to the reference model.
Upper two rows represent the map view of the synthetic model (left) and the reconstructed anomalies
(right) at 0 and 5 km depth. Location of the profile is shown. Lower plots represent the synthetic model and
reconstruction result in the cross section.
4. Synthetic modeling
4.1. General remarks
The ATOM-3D software provides a wide range of possibilities for performing various
synthetic tests. In the actual version there is a possibility to define the initial model using
four different ways:
1. checkerboard;
2. arbitrary anomalies defined in map view (horizontal anomalies);
3. arbitrary anomalies defined in some vertical sections (vertical anomalies);
4. checkerboard defined along vertical sections;
The travel times for the synthetic test are computed by tracing of rays between the
sources and receivers corresponding to the real observation system. As in case of real
data processing, bending algorithm is used.
Organization of folders for all kinds of synthetic models is generally the same as in case
of the real data model. The Model folder name should consist of 8 characters and have
the same structure of subfolders as the real data models. However, in the “ini_data”
folder, two additional files should be added:
1. “anomaly.dat” which contains the description of the synthetic model. First line in this
file define the way of definition of the model (1, 2, 3, or 4). The following description
depends on the case and will be described in details in the corresponding sections.
2. “noise.dat” which defines the shape and amplitude of noise added to the synthetic data
(see the example below).
4.2. Definition of noise
The noise is produced by generator of random numbers with a realistic histogram. An
example:
\DATA\DATASET1\BRD_NSE2\ini_param\noise.dat
1 type of noise 1: random, 2: from real data
______________________________________
0.15 0.25
level of noise for P and S data
0
in percents, partition of outliers
10.
times larger than the normal noise
0. Amplitude of error at the stations
**************************************
Histogram of noise distridution:
1000
nhist number of nodes for histogram description
9.9999997E-05
-9.875000
5.5515502E-02
-9.625000
6.5609232E-02
-9.375000
5.8399424E-02
-9.125000
7.6423936E-02
:::::::::::::::::::::::::::::::
7.875000
8.125000
8.375000
8.625000
0.1441961
0.1564528
0.1319394
0.1333814
8.875000
9.125000
9.375000
9.625000
9.875000
10.12500
0.1268926
0.1348234
0.1189618
0.1002163
0.1189618
0.0000000E+00
In this example standard deviation of noise is 0.15 s for P data and 0.3 s for S data
(actually, S data are not used). In addition, the algorithm allows adding the outliers to test
the robustness of the solution in case of some rough errors of phase picking. In the
presented case, no outliers are added.
The basic shape of the noise distribution histogram is given in this file. In this case, the
histogram is described by 1000 nodes and is distributed from -10 to 10 s. This histogram
corresponds to the distribution of real residuals in the ISC catalogue.
4.3. Visualization of the initial synthetic model in horizontal and
vertical sections
It is recommended to control the correctness of the definition using the programs in
Projects:
\PROG\a_set_syn_hor for horizontal presentations
\PROG\a_set_syn_ver for vertical presentations
The names of area and models are defined in both cases in the same directory in the file
“SET.DAT”. The depths and limits of the horizontal sections for visualization are defined
in \DATA\DATASET1\board_01\ini_param\sethor.dat, the same as used
for visualization of real data horizontal sections (Section 3.1). The coordinates of the
cross-section and vertical limits for visualization are defined in
\DATA\COSTARIC\BRD_VER1\ini_param\setver.dat, the same as used for
visualization of real data vertical sections (Section 3.2).
The output of horizontal presentation is as the same as used for presenting real data
results:
/FIG_files/hor/hor_M.grd: relative anomalies in percent in horizontal sections.
M is a number of depth levels according to the information in “sethor.dat“.
The output of vertical presentation is written to:
/FIG_files/syn_ini/ver_M.grd: relative anomalies in percent in horizontal
sections. M is a number of cross sections according to the information in “setver.dat“.
These files can be directly visualized in the Surfer Software as a contour line plot.
In a case of rather complicated definition of horizontal or vertical anomalies, it is
recommended to perform both Projects a_set_syn_ver and a_set_syn_hor .
4.4. Definition of the checkerboard anomalies
In case of a regular checkerboard model, the indicator in the first line of “anomaly.dat”
file should be 1. The following lines contain description of the checkerboard. An example
is presented below:
ATOM_3D\DATA\DATASET1\BOARD_01\ini_param\anomaly.dat
1
1 - board, 2 - horiz. anom, 3 - vert. anom, 4 – vert. board
_______________________________________________
7.00 anom_P: P-anomalies
-30. 30. 5. 0.0 xmin_P, xmax_P, dx_anom_P, dx_empty_P: BOARD along
-30. 30. 5. 0.0 ymin_P, ymax_P, dy_anom_P, dy_empty_P: BOARD along
-3. 200. 5. 0.0 zmin_P, zmax_P, dz_anom_P, dz_empty_P: BOARD along
7.00 anom_S: S-anomalies
-30. 30. 5. 0.0 xmin_S, xmax_S, dx_anom_S, dx_empty_S: BOARD along
-30. 30. 5. 0.0 ymin_S, ymax_S, dy_anom_S, dy_empty_S: BOARD along
-3. 200. 5. 0.0 zmin_S, zmax_S, dz_anom_S, dz_empty_S: BOARD along
X direction
Y direction
Z direction
X direction
Y direction
Z direction
In the presented example both P and S anomalies are +_7% amplitude (actually, S
anomalies are not used). The size of anomalies is 5 km (e.g. dx_anom_P=5). There is a
possibility to define empty space between anomalies (dx_empty_P). Along Z direction,
the anomaly is the same down to 200 km depth. It is important to define the anomaly not
from a zero depth. In this case, if the anomaly is negative, the algorithm of ray tracing
will try to bend the ray upward to achieve the minimum of travel time. In the presented
example the anomalies are defined from -3 km depth.
A model corresponding to the presented example is shown in figure 4.1. In the same
figure we present also the results of its reconstruction
Figure 4.1. Checkerboard test. Upper left map represents the initial synthetic distribution. The other plots
are the results of reconstruction at different depths after five iterations.
This way allows also definition of horizontal blocks with unchanged values along X or Y.
It is also possible to define single anomalies.
4.5. Definition of free horizontal anomalies
In case of synthetic model definition using free horizontal anomalies, the indicator in the
first line of “anomaly.dat” file should be 2. In this case, a new subfolder “forms” should
be created within the model folder. This folder contains the files with descriptions of the
shapes which can be used for definition of the model. The names of these files should
consist of any five characters and have the extension “.bln’. It can be simple forms (e.g.
triangle, square, circle etc) or more complicated shapes. In practice, the synthetic
anomalies can be created according to the shapes of patterns observed in the real data
inversion. In this case, it is recommended to use a tool “map/digitize” in Surfer Software
to produce the shapes in geographical coordinates which correspond to the really
observed features. The curve which determines the anomaly should be not necessarily
closed. Example of such a file generated in Surfer is presented below.
Example of
ATOM-3D\\DATA\DATASET1\LOTOS_01\forms\smile.bln
20,1
-71.057525936,
-71.023617952,
-70.981234496,
-70.930373536,
-70.879512576,
-70.828651616,
-70.777790656,
-70.735406184,
-70.693022728,
-70.709975704,
-70.743882672,
-70.782028392,
-70.820174112,
-70.862558584,
-70.904943056,
-70.947326512,
-70.981234496,
-71.002426224,
-71.023617952,
-71.040571944,
-21.128906016
-21.154336496
-21.171290488
-21.184005728
-21.18824448
-21.192482216
-21.192482216
-21.18824448
-21.171290488
-21.222151448
-21.25182068
-21.273012408
-21.27725116
-21.285727648
-21.281488896
-21.268773656
-21.247581928
-21.2263902
-21.196720968
-21.167051736
Information about the synthetic anomaly is presented in the file “anomaly.dat“. An
example is presented below:
Example of
ATOM-3D\DATA\DATASET1\LOTOS_01\ini_param\anom.dat
2
1 - board, 2 - horiz. anom, 3 - vert. anom, 4 – vert. board
_______________________________________________
11
number of anomalies
*******************************
L1____
Figure
0. 0. 0. 0.
-8 -8
-50. 150.
*******************************
O1out
Figure
0. 0. 0. 0.
8 8
-50. 150.
*******************************
O1_in
Figure
0. 0. 0. 0.
-8 -8
-50. 150.
*******************************
T1____
Figure
0. 0. 0. 0.
-8 -8
-50. 150.
*******************************
O2out
Figure
0. 0. 0. 0.
8 8
-50. 50.
*******************************
O2_in
Figure
0. 0. 0. 0.
-8 -8
-50. 50.
*******************************
S1____
Figure
0. 0. 0. 0.
-8 -8
-50. 50.
*******************************
circl
Figure
0. 0. 0. 0.
4 4
-50. 50.
*******************************
glaz1
Figure
0. 0. 0. 0.
-8 -8
-50. 50.
*******************************
glaz2
Figure
0. 0. 0. 0.
-8 -8
-50. 50.
*******************************
smile
Figure
0. 0. 0. 0.
-8 -8
-50. 50.
In this example the synthetic model consists of 11 horizontal prisms. All of them are
located in the depth interval between -50 and 50 km. All of them should be defined in
folder “forms” in files “L1___.bln“, “Oout1.bln“, “O_in1.bln“, “T1___.bln“ etc. It is
important to define the anomaly not from a zero depth. In this case, if the anomaly is
negative, the algorithm of ray tracing will try to bend the ray upward to achieve the
minimum of travel time. In the presented example the anomalies are defined from -10 km
depth.
In addition, the entire image can be scaled and rotated. To do this, use the file:
ATOM-3D\\DATA\DATASET1\LOTOS_01\forms\scaling. dat
-16.8 -16.4
28.1 28.35
270
In this example is defined in the interval of longitude of -16.8° - -16.4° and latitude 28.1°
- 28.35° and is rotated to the angle of 45°
If this file is not defined, the model would be created according to coordinates defined in
forms.
A model corresponding to the presented example is shown in figure 4.2. In the same
figure we present also the results of its reconstruction in P and S data.
Figure 4.2. Synthetic test with free horizontal anomalies according to the model ATOM3D\\DATA\DATASET1\LOTOS_01. Upper left map represents the initial synthetic distribution. The
other maps are the results of reconstruction at different depths.
4.6. Definition of free vertical anomalies
There is a possibility to define the synthetic model as superposition of prisms which have
fixed shape in some vertical sections. In this case, the indicator in the first line of
“anomaly.dat” file should be 3. A new subfolder “forms” should be created within the
model folder. This folder contains the files with descriptions of the shapes which can be
used for definition of the model. The names of these files should consist of any five
characters and have the extension “.bln’. It can be simple forms (e.g. triangle, square,
circle etc) or more complicated shapes. In practice, the synthetic anomalies can be
created according to the shapes of patterns observed in the vertical sections of the real
data inversion results. In this case, it is recommended to use a tool “map/digitize” in
Surfer Software to produce the shapes in geographical coordinates which correspond to
the really observed features. The curve which determines the anomaly should be not
necessarily closed. Example of such a file generated in Surfer is presented below.
Example of
\DATA\DATASET2\VER_free\forms\smile.bln
20,1
-71.057525936,
-71.023617952,
-70.981234496,
-70.930373536,
-70.879512576,
-70.828651616,
-70.777790656,
-70.735406184,
-70.693022728,
-70.709975704,
-70.743882672,
-70.782028392,
-70.820174112,
-70.862558584,
-70.904943056,
-70.947326512,
-70.981234496,
-71.002426224,
-71.023617952,
-71.040571944,
-21.128906016
-21.154336496
-21.171290488
-21.184005728
-21.18824448
-21.192482216
-21.192482216
-21.18824448
-21.171290488
-21.222151448
-21.25182068
-21.273012408
-21.27725116
-21.285727648
-21.281488896
-21.268773656
-21.247581928
-21.2263902
-21.196720968
-21.167051736
Information about the synthetic anomaly is presented in the file “anomaly“. An example
is presented below:
Example of
\DATA\DATASET2\VER_free\ini_param\anom.dat
3
1 - board, 2 - horiz. anom, 3 - vert. anom
_______________________________________________
7
number of anomalies
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
L1____
Figure
0. 0. 0. 0.
-8 -8
-150. 150.
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
O1out Figure
0. 0. 0. 0.
8 8
-150. 150.
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
O1_in Figure
0. 0. 0. 0.
-8 -8
-150. 150.
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
T1____
Figure
0. 0. 0. 0.
-8 -8
-150. 150.
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
O2out Figure
0. 0. 0. 0.
8 8
-150. 150.
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
O2_in Figure
0. 0. 0. 0.
-8 -8
-150. 150.
*******************************
-70.82538797 -22.55326057 -71.14119379 -18.15829857
S1____
Figure
0. 0. 0. 0.
-8 -8
-150. 150.
In this example the synthetic model consists of seven prisms defined in one cross section.
In fact, any amount of prisms can be defined in any different cross sections at the same
time. All of them should be defined in folder “forms” in files with corresponding names
In addition, the entire image can be scaled and rotated. To do this, use the file:
\DATA\DATASET2\VER_free\forms\scaling.dat
120 380
10 70
0
In this example is defined in the interval of X of 120 - 380 and Z of 10 - 70 without
rotation
If this file is not defined, the model would be created according to coordinates defined in
forms.
A model corresponding to the presented example is shown in figure 4.3.
0
-20
-40
-60
-80
0
50
100
150
200
250
300
350
400
450
Figure 4.3. Synthetic model defined by free vertical patterns.
4.7. Definition of vertical checkerboard anomalies
For investigating vertical resolution we can defines checkerboard model at vertical
sections. In this case, the indicator in the first line of “anomaly.dat” file should be 4. The
following lines contain description of the checkerboard. An example is presented below:
ATOM_3D\DATA\DATASET1\VBRDmod2\ini_param\anomaly.dat
4
1 - board, 2 - horiz. anom, 3 - vert. anom
_______________________________________________
1 number of profiles
***********************************
-17 28.21 -16.3 28.21 Fi_A,Tet_A,Fi_B,Tet_B: coordinates of ends of the profile A-B
-20 20
y_right,y_left: thickness of the anomalies along Y
7.00
amplitude of anomalies
0. 100. 4. 0.0 xmin_P, xmax_P, dx_anom_P, dx_empty_P: BOARD along X direction
-5. 9. 7. 0.0 zmin_P, zmax_P, dz_anom_P, dz_empty_P: BOARD along Z direction
***********************************
In the presented example the anomalies are +_7% amplitude. The size of anomalies is 4
km along X and 7 km along Z. There is a possibility to define empty space between
anomalies. These anomalies are defined across the section with the ends defined in:
-17 28.21 -16.3 28.21 coordinates of the profile
Thickness of blocks across the profile is from -90 km to 90 km.
.
A model corresponding to the presented example is shown in figure 3.2 in Section for
visualizing results.
4.8. Practical realization of ATOM-3D code for synthetic tests
To perform the successful run of the ATOM-3D for synthetic tests, the data structure
should be created, and the anomalies should be defined, as described in section above.
There is possibility to run the steps of synthetic data calculation and inversion manually,
step by step. However, the ATOM-3D code contains a program which performs automatic
managing of all steps. The source of this program is presented below:
Program for automatic managing of the ATOM-3D synthetic modeling steps:
File “ATOM_3D\PROG\START_synth_inversion\start_syn.f90”
(the program steps are highlighted with red)
USE DFPORT
character*8 ar,ar_all(10),md,md_all(10),line
character*1 rg_all(100),rg,it
integer kod_loc(10),kod_iter(10),kod_oe(10)
open(1, file='../../all_areas.dat')
do i=1,4
read(1,*)
end do
do i=1,10
read(1,'(a8,1x,a8,1x,i1,1x,i1,1x,i1)',end=7)ar_all(i),md_all(i),kod_iter(i),kod_oe
(i)
write(*,*)ar_all(i),md_all(i),kod_iter(i),kod_oe(i)
end do
7 close(1)
n_ar=i-1
! Check all the areas:
do iar=1,n_ar
ar=ar_all(iar)
md=md_all(iar)
niter=kod_iter(iar)
koe=kod_oe(iar)
open(11,file='../../model.dat')
write(11,'(a8)')ar
write(11,'(a8)')md
write(11,'(i1)')1
write(11,'(i1)')1
write(11,'(i1)')koe
write(11,'(i1)')1
close(11)
!Compute the synthetic times:
***********************************
i=system('..\b_synth_times\rays.exe')
!******************************************************************
open(1,file='../../data/'//ar//'/'//md//'/ini_param/MAJOR_PARAM.DAT')
do i=1,10000
read(1,'(a8)',end=573)line
if(line.eq.'ORIENTAT') goto 574
end do
573 continue
write(*,*)' cannot find ORIENTATIONS in MAJOR_PARAM.DAT!!!'
pause
574 read(1,*)nornt
close(1)
! Execute the inversion for grids with different orientations:
do iter=1,niter
write(it,'(i1)')iter
open(11,file='../../model.dat')
write(11,'(a8)')ar
write(11,'(a8)')md
write(11,'(i1)')iter
write(11,'(i1)')1
write(11,'(i1)')koe
write(11,'(i1)')1
close(11)
write(*,*)'
****************************************************'
write(*,*)'
Residuals for active data:'
i=system('..\1a_resid\resid.exe')
999
continue
do igr=1,nornt
open(11,file='../../model.dat')
write(11,'(a8)')ar
write(11,'(a8)')md
write(11,'(i1)')iter
write(11,'(i1)')igr
close(11)
if(iter.eq.1) then
write(*,*)'
****************************************************'
write(*,*)'
Compute the ray density'
i=system('..\2_ray_density\plotray.exe')
write(*,*)'
****************************************************'
write(*,*)'
Compute the parameterization grid:'
i=system('..\3_grid\grid.exe')
i=system('..\4_tetrad\Tetrad.exe')
i=system('..\5_sosedi\add_matr.exe')
end if
i=system('..\6_matr\matr.exe')
i=system('..\7_invers\Invbig.exe')
end do
i=system('..\8_3D_model\mod_3D.exe')
end do ! Different grids
end do ! Different areas
stop
end
This program allows running all the ATOM-3D steps for one or several models. The list
of models is defined in file ATOM_3D/all_areas.dat in a same way as when real data
inversion was performed (Section 2.4).
File “ATOM_3D\all_areas.dat”
1-2: codes of the area and models (each contain 8 characters)
3: number of iterations
4: 0 all data, 1 odd events, 2 even events
**********************************************
DATASET1 BOARD_01 5 0
DATASET1 LOTOS_01 5 0
DATASET1 BRD_VER1 5 0
In the presented example, three synthetic models are defined for the same area
“DATASET1“: BOARD_01, LOTOS_01, and BRD_VER1. The configuration of the
observation systems for these tests are taken from the file “ATOM3D/inidata/rays.dat”. The third number in of every second line is number of the
performed iterations, which is always 5, in our example. The last index is not used in this
case.
It is important to define all the parameters in the file “all_areas.dat” according to a fixed
format. They will run consequently one after another.