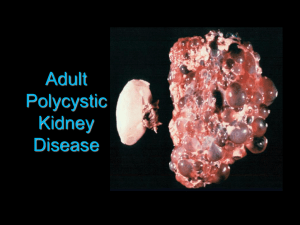
the renal gene ontology annotation (goa) initiative
advertisement

S16(W) THE RENAL GENE ONTOLOGY ANNOTATION INITIATIVE. Alam-Faruque, Y1, Dimmer, E1, Huntley, R1, Foulger, R1, Tweedie, S2, Hill, D3, Harris, M1, O’Donovan, C1, Scambler, P4, Apweiler, R1. 1 EMBL-European Bioinformatics Institute, The Wellcome Trust Genome Campus, Hinxton, Cambridgeshire, 2FlyBase, Department of Genetics, University of Cambridge, 3 The Jackson Laboratory , Bar Harbor, ME, USA; 4UCL-Institute of Child Health, London The Gene Ontology (GO) is the established standard for the annotation of gene products that describes their molecular functions, biological processes and subcellular locations. The Renal GO Annotation Initiative (part of the GOA project in the UniProtKB group) aims to provide high quality, detailed GO annotation for mammalian proteins implicated in renal development and function. With the recent technological advancement enabling renal researchers to generated high-quality, large-scale and valuable experimental data, the use of GO to curate scientific literature will ensure that by integrating results from these peer-reviewed experiments, the accumulated knowledge of renal development and function can be exploited to fully benefit researchers seeking to understand and alleviate renal diseases. Currently, ~1730 gene products have been identified as targets involved in renal function and development. There was previously a lack of comprehensive GO terms describing renal-specific processes therefore, in close collaboration with renal experts, the project has recently improved the Gene Ontology for describing the complex process of renal development. The number of GO terms has been increased from 22 to 468 with the creation of an extra 446 new terms that detail not only the anatomical description but also the biological processes that contribute to the development of the renal system. The new GO terms are applicable to development of the renal system across various species, including the pronephros (amphibia), mesonephros (fish), metanephros (mammalia) and Malpighian tubule (insecta). Various gene products have been annotated to some of these new renal development terms and hence the renal community can use the high quality, detailed annotations and the specific GO terms to improve identification of renal development pathways and processes that large gene sets may be involved in and hence generate new biological insights into renal diseases. An overview of the aims, benefits and progress of the Renal GO Annotation Initiative to date will be outlined at the 2011 BRS/RA Conference. The project very much welcomes any feedback from the renal research community, including suggestions on additional genes/proteins or datasets which users would like to see curated, or where the descriptiveness of the Gene Ontology for any aspect of renal biology could be improved. Further information on this project is available at http://www.geneontology.org/GO.renal and http://www.ebi.ac.uk/GOA/kidney/ .