The top three proteins with the most similarity to the nitrite reductase
advertisement

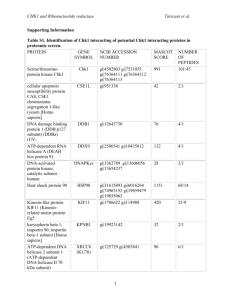
Nitrate assimilation related genes sequence identification and multiple alignments: The top three proteins with the most similarity to the nitrite reductase [NAD(P)H] large subunit nasD were from the Bacillus megaterium DSM319 (YP_003596351) with 99% identity, Bacillus megaterium WSH-002 (YP_005496278) with 99% identity, Bacillus megaterium QM B1551 (YP_003561620) with 99% identity. The nitrite reductase [NAD(P)H] large subunit nasD of Bacillus megaterium NCT-2 shared high similarity with the proteins from the three Bacillus megaterium species. Refer to Fig. 1 for detailed multiple alignments results. Fig. 1 Multiple alignments of nitrite reductase [NAD(P)H] large subunit from similar species Bacillus megaterium NCT-2: AHTF00000000; Bacillus megaterium DSM 319: YP_003596351; Bacillus megaterium WSH-002: YP_005496278; Bacillus megaterium QM B1551: YP_003561620. Residue with no consensus or the residue is neither identical nor similar to the consensus was printed in a normal rendition, residue that is identical to the column-consensus was printed in an inverse rendition, residue that is not identical but at least similar to the column-consensus was printed in a gray background rendition. The consensus column was marked with “*”, non-consensus column was marked with “.”. The top three proteins with the most similarity to the nitrite reductase [NAD(P)H] small subunit nasE were from the Bacillus megaterium QM B1551 (YP_003561621) with 100% identity, Bacillus sp.1NLA3E (ZP_09600724) with 72% identity, Bacillus atrophaeus 1942 (YP_003975907) with 70% identity. The nitrite reductase [NAD(P)H] small subunit nasE of Bacillus megaterium NCT-2 shared high similarity with the proteins from one Bacillus megaterium species and medium similarities with other two Bacillus species. Refer to Fig. 2 for detailed multiple alignments results. Fig. 2 Multiple alignments of nitrite reductase [NAD(P)H] small subunit from similar species Bacillus megaterium NCT-2: AHTF00000000; Bacillus megaterium QM B1551: YP_003561621; Bacillus sp.1NLA3E: ZP_09600724; Bacillus atrophaeus 1942: YP_003975907. Residue with no consensus or the residue is neither identical nor similar to the consensus was printed in a normal rendition, residue that is identical to the column-consensus was printed in an inverse rendition, residue that is not identical but at least similar to the column-consensus was printed in a gray background rendition. The consensus column was marked with “*”, non-consensus column was marked with “.”. The top three proteins with the most similarity to the glutamine synthetase glnA were from the Bacillus megaterium DSM 319 (YP_003599266) with 100% identity, Bacillus megaterium QM B1551 (YP_003564545) with 99% identity, Bacillus subtilis subsp.spizizenii ATCC 6633 (ZP_06873437) with 89% identity. The glutamine synthetase glnA of Bacillus megaterium NCT-2 shared high similarity with the proteins from the two Bacillus megaterium species and with relatively high similarity with the Bacillus subtilis species. Refer to Fig. 3 for detailed multiple alignments results. Fig. 3 Multiple alignments of glutamine synthetase from similar species Bacillus megaterium NCT-2: AHTF00000000; Bacillus megaterium DSM 319: YP_003599266; Bacillus megaterium QM B1551: YP_003564545; Bacillus subtilis subsp.spizizenii ATCC 6633: ZP_06873437. Residue with no consensus or the residue is neither identical nor similar to the consensus was printed in a normal rendition, residue that is identical to the column-consensus was printed in an inverse rendition, residue that is not identical but at least similar to the column-consensus was printed in a gray background rendition. The consensus column was marked with “*”, non-consensus column was marked with “.”. 3D structure prediction: The nitrite reductase also consists of two components: the nitrite reductase [NAD(P)H] large subunit (nasD) and the nitrite reductase [NAD(P)H] small subunit (nasE). The nitrite reductase [NAD(P)H] large subunit nasD predicted by SWISS MODEL (Fig. 4a) was modeled base on the sequence residues 144-183, this modeling range shared a 37.5% similarity to the modeling template 2XOL A (3.00 Å). While the nitrite reductase [NAD(P)H] large subunit nasD shared a 32.0% similarity to the template protein 3KLJ A (2.10 Å) (Fig. 4b) predicted by NCBI blastp with query coverage of 48.0%. The nitrite reductase [NAD(P)H] small subunit nasE predicted by SWISS MODEL (Fig. 4c) was modeled base on the sequence residues 5-107, this modeling range shared a 29.1% similarity to the modeling template 2ZYL A (2.30 Å). While the nitrite reductase [NAD(P)H] small subunit nasE shared a 40.0% similarity to the template protein 2ZYL A (2.30 Å) (Fig. 4d) predicted by NCBI blastp with query coverage of 60.0%. Also, due to the low homology to the modeling template, the nitrite reductase [NAD(P)H] large subunit nasD predicted by SWISS MODEL was only partially modeled. Thus it only displayed a partially 3D structure compared with its most similar known protein from the PDB database by NCBI blastp. While the 3D structure of nitrite reductase [NAD(P)H] small subunit nasE predicted by SWISS MODEL was highly identical to that of the protein model from the PDB database by NCBI blastp. a b c d Fig. 4 3D structure modeling of nitrite reductase a: nasD predicted by SWISS MODEL; b: nasD most similar protein predicted by NCBI blastp; c: nasE predicted by SWISS MODEL; d: nasE most similar protein predicted by NCBI blastp. The glutamine synthetase is designated as glnA. The glutamine synthetase glnA predicted by SWISS MODEL (Fig. 5a) was modeled base on the sequence residues 5-443, this modeling range shared a 38.9% similarity to the modeling template 1F52 E (2.49 Å). While the glutamine synthetase glnA shared a 41.0% similarity to the template protein 2GLS A (3.50 Å) (Fig. 5b) predicted by NCBI blastp with query coverage of 98.0%. It showed that the 3D structure of glutamine synthetase glnA predicted by SWISS MODEL was highly identical to that of the protein model from the PDB database by NCBI blastp. a b Fig. 5 3D structure modeling of glutamine synthetase a: glnA predicted by SWISS MODEL; b: glnA most similar protein predicted by NCBI blastp.