Current Research in Microbiology and Biotechnology
Vol. 1, No. 4 (2013): 173-182
Research Article
Open Access
ISSN: 2320-2246
Isolation and molecular characterization of Bacillus
megaterium isolated from various agro climatic
zones of Karnataka and its effect on medicinal plant
Ruta gradiolus
HS Ravikumar Patil1*, T Vasantha Naik2, BR Vijay Avin3 and HA Sayeswara4
Department of Biotechnology, G.M. Institute of Technology, Davangere-577066, Karnataka, India
Department of Botany, D.R.M.Science College, Davangere-577066, Karnataka, India
3 Department P.G.Studies and Research in Biotechnology, Sahyadri Science College (Autonomous), Kuvempu University,
Shivamogga-577203, Karnataka, India
4 Department of Zoology, Sahyadri Science College (Autonomous), Kuvempu University, Shivamogga-577203, Karnataka, India
1
2
* Corresponding author: HS Ravikumar Patil, email: patil_varuni@gmail.com
Received: 08 June 2013
Accepted: 23 June 2013
Online: 01 July 2013
ABSTRACT
Bacillus megaterium strains from different agro climate zones will be isolated, identified and confirmed using
microscopic observation, biochemical and physiological characters like Gelatin Liquefaction, Levan formation
test, Indole production test and catalase test. The identified pure cultures of these isolates will be maintained for
further studies. Molecular diversity of these isolates will be characterized by RAPD marker analysis. The RAPD
banding pattern of these isolates is used to distinguish the isolates of the different zones. Simultaneously plants
growth response studies using Ruta gradiolus as host was conducted. In plants inoculated with Bacillus
megaterium, the initial height, number of branches, fresh and dry weight of the roots and shoots, will be noted
and compared with the control (Uninnoculated plants). Biochemical parameters like chlorophyll estimation will
be detected and compared with the control and surprisingly, wide diversity of these isolates were observed in
RAPD pattern.
Keywords: Bacillus megaterium, agro climatic zones, Ruta gradiolus, RAPD
INTRODUCTION
Microorganisms are present all most everywhere. They
are ubiquitous and at the same time highly
proliferating. Modern agriculture, apart from
improving the overall production and productivity, has
also caused destruction to the environment. The use of
chemical fertilizers was necessitated by cultivation of
high yielding varieties which has resulted in
degradation of soil health [1]. Hence, alternative ecofriendly methods for sustainable agriculture are being
envisaged. Biological methods offer an excellent
alternate strategy for effective control of various
diseases and augmentation of nutrient availability in
the rhizosphere.
growth [2]. They are rhizosphere bacteria that can
benefit plant growth by different mechanisms [3].
Recent progress in the understanding of their diversity,
colonization ability, mechanisms of action, formulation,
and application should facilitate their development as
reliable components in the management of sustainable
agricultural systems. Microorganisms are involved in a
range of processes that affect the transformation of
different soil nutrients such as Nitrogen, Phosphorous
etc and thus are integral part of the soil nutrient cycle.
Plant growth promoting Rhizobacteria (PGPR) are soil
bacteria that colonize the roots of plants following
inoculation onto seeds or soil and that enhance plant
A plant even possess active defense mechanisms
become infected by a virulent pathogen, because the
pathogen avoids triggering or suppress resistance
reaction or evades the effect of activated defenses. If
defense mechanisms are triggered by a stimulus prior
to infection by a plant pathogen, disease can be
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
173
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
reduced. Induced resistance is a state of enhanced
defensive capacity developed by a plant appropriately
stimulated by certain chemicals non-pathogens, a
virulent form of pathogens; incompatible races of
pathogens or by virulent pathogens under
circumstance were infection is stalled owing to
environmental conditions [4].
A massive accumulation of phytoalexin, phenolic
compounds [5], increase in the activities of PR proteins
[6], peroxidase [7], increase in the levels of mRNA’s
encoding phenyl alanine ammonia lyase (PAL) and
enhanced lignifications have been reported in plants
following treatments with PGPR strains.
The effects of PGPR on plant growth can be mediated
by the direct or indirect mechanisms. The direct effects
have been most commonly attributed to the production
of plant hormones such as auxins, gibberllins and
cytokines as by supplying biologically fixed nitrogen.
These PGPR also affect growth of plants by indirect
mechanisms such as suppression of bacterial and
fungal, nematode pathogens by production of
antibiotics, siderophores, ammonia etc., by induced
systemic resistance and or by competing with
pathogens for nutrients [8].
Molecular analysis of genomic DNA of the organism is
useful for distinguishing the bacterial strains better at
interspecies level these techniques provide valuable
information on the magnitude of genetic variability
within and between organisms of different species.
With the advent of molecular techniques, several
arbitrary primers based Randomly Amplified
Polymorphic DNA (RAPD) technique has been used for
typing and identification of number of closely related
species of bacteria and assessment of genetic
relationships. Its results are usually consistent with
those of DNA-DNA homology studies and can be used to
estimate the genetic distances [9].
Table 1. Characteristic features of agro climatic zones of Karnataka
Zone
1
2
3
4
5
6
7
8
9
10
Name of
the
Zones
North
eastern
transition
zone
North
eastern
dry zone
North dry
zone
Central
dry zone
Eastern
dry zone
Southern
dry zone
Southern
transition
zone
Northern
transition
zone
Hilly
zone
Coastal
zone
Soil Type
Rainfall
(mm/y)
Temperature
(o C)
Humidity
(%)
Sand
(%)
Silt
(%)
Clay
(%)
pH
Laterite Soil
860
31.1-20.7
65.0
55.7
10.1
34.2
8.07
CEC
(Centi
moles)
30.00
Sandy Clay
Loam
526754
22.3-33.5
63.0
14.3
24.7
61.0
8.5
63.00
Clay Loam
585
32.4-21.9
67.0
17.86
17.98
64.16
9.17
44.68
Red Sandy
loam to
Black soil
Red sandy
soil
Red sandy
soil
Red sandy
loam
456717
30.8-20.7
73.0
34.66
28.96
36.38
9.54
47.82
645889
670.6888.6
6191300
29.2-18.6
71.0
72.97
4.93
22.1
5.31-6.21
16.5
29.1(max)
61.6
71.45
4.62
23.93
6.72
15.5
30-19.2
81.0
76.17
5.86
17.94
5.8
-
Black soil
780
30.1-18.0
76.0
16.35
36.38
57.27
6.72-7.85
64.0
Red loam to
red sandy
loam
Lateritic
9043695
25.2-16.6
89.0
69.7
20.5
9.79
5.3
20.5
4000
30.5-23.5
96.5
48.46
12.76
38.78
5.2
25.5
*Source: department of soil Science, UAS, GKVK, Bangalore
The present investigation was carried out to isolate and
identify Bacillus megaterium from soil of different agro
climatic zones of Karnataka, to characterize B.
megaterium using RAPD markers was done in
Department
of
Biotechnology,
University
of
Agricultural Sciences, G.K.V.K Campus, Bangalore, India.
soil type, cropping pattern and other climatic
conditions. In order to study the biological and
molecular variability in Bacillus megaterium in the soils
of different zones of Karnataka, soil samples were
collected from each agro climatic zone. Four sampling
sites were selected randomly for each agro climatic
zone.
Collection of soil samples from different agro
climatic zones of Karnataka
The geographical area of Karnataka state is divided into
ten agro climatic zones on the basis of annual rainfall,
Soil sampling
Four soil samples of 500 grams each were collected
randomly from top six-inch layer of soil from each and
packed in polyethylene bags.
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
174
MATERIALS AND METHODS
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
Isolation of Bacillus megaterium from different
zones
Bacillus megaterium was isolated from the soils
collected from different zones, by growing in glucose
mineral agar media .For isolation, cell material was
checked microscopically for the presence of typical
cells of Bacillus megaterium and purified on nutrient
agar. Bacillus megaterium is a common inhabitant of the
soil. As characteristic of many soil bacteria, Bacillus
megaterium form
endospores. An endospore is a
resting structure formed inside some bacteria, which
enable the bacterium to survive harsh environmental
conditions such as heat and drying. The cell wall of
Bacillus megaterium is covered by a sticky
polysaccharide called capsule which causes the
individual cells of the bacterium to stick together in
chains. Bacillus megaterium was isolated and dilutions
were made upto 10-3 and 10-4. Then plated on glucose
mineral base agar medium and incubated at 300C for 2
days. Thereafter all the isolates were subjected to
various tests for confirming their identity.
Identification of Bacillus megaterium
Colony morphology and microscopic examination
All the isolates and standard strains formed completely
white, round, smooth and shiny colonies. During
microscopic examination all the isolates were found to
be gram positive rods. Presence of endospores was
confirmed by endospore staining.
Physiological tests for Bacillus megaterium
All the physiological tests mentioned were conducted
in duplicate for each isolate.
Gelatin liquifaction
The method followed was as described earlier [10], in
brief, Petriplates containing gelatin agar were spotted
with overnight grown bacterial culture at 300C and
incubated for 3 days. The plates were then flooded with
12% HgCl2 solution and allowed to stand for 20
minutes and observed for clear zones around the
growth of bacterium which indicated gelatin
liquifaction.
Acid gas production
This method followed was as described earlier [11], in
brief Bacterial isolates were tested for acid and gas
production by inoculating 5 ml of the sterile glucose
broth with bromocresol purple (15 ml/l of 0.04%
solution as pH indicator) in test tubes containing
Durham’s tube. The tubes were incubated for seven
days at 300C. Accumulation of gas in these Durham’s
tube was taken positive for gas production and change
in colour of the medium to yellow was taken as positive
for acid production.
Catalase test
Nutrient slants were incubated at 300C for 24 hrs. After
incubation these tubes were flooded with 1ml of 3%
H2O2 and observed for gas bubbles. Occurrence of gas
bubbles was scored +ve for catalase test.
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
Pot Experiment
Inoculum preparation
Bacillus megaterium isolates were grown separately, in
a 250 ml flask containing 100ml NA for 2 days. The
grown cultures were homogenized and 40 ml of each of
the solution were inoculated to each pot.
Treatment allocation
T0 - Control (uninoculated control)
The three varieties of aerobic rice selected for the study
were MAS 109, MAS 26 and MAS 946. Three
replications were done for each treatment with one
plant per pot. Each pot with 4kg of soil. The total
sample size for recording the observation was 99
plants, along with the control. No external supply of
nutrients was done for the experimental plants.
Effect of Bacillus megaterium on aerobic rice (Oryza
sativa)
Plant growth parameters
The observations with respect to the growth
parameters including plant height, number of leaves,
number of tillers, shoot fresh weight, shoot dry weight,
root fresh weight, root dry weight, and N, P, chlorophyll
content and total sugar concentration were recorded.
Plant height
The plant height was measured from the soil surface to
the tip of the growing point at 30, 60, 90 Days after
Planting and at maturity.
Number of leaves
The numbers of fully opened leaves were recorded at
30, 60, 90 Days after planting and at maturity.
Number of tillers
Number of tillers were recorded at 30, 60, 90 DAP and
at maturity.
Number of panicles
Number of panicles was recorded after maturity.
Plant Biomass
Root fresh weight
The harvested plants were weighed and then the root
fresh weight was recorded and expressed as grams per
plant.
Root dry weight
The harvested roots were dried in an oven at 600C for 2
days to attain constant weight and then the root dry
weight was recorded and expressed as grams per plant.
Shoot fresh weight
The harvested plants were weighed and then the shoot
fresh weight was recorded and expressed as grams per
plant.
Shoot dry weight
175
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
The harvested plants were dried in an oven at 600C for
4 days to attain constant weight and then the dry
weight was recorded and expressed as grams per plant.
Grain weight
The grain weight per plant was recorded after harvest
and expressed as grams per plant.
Biochemical studies of plants inoculated with
Bacillus megaterium
Estimation of Chlorophyll content of leaves
Chlorophyll content in Rice leaves was determined by
the following procedure described earlier [12]. One
hundred milligram of leaf tissue was placed in a vial
containing 7 ml of DMSO and chlorophyll was extracted
in to the fluid by incubating at 650C overnight. The
extract was then transferred to a graduated tube and
made up to a total volume of 10 ml with DMSO, assayed
immediately or transferred to vials and stored between
0 – 40C until required for analysis.
Assay was done by transferring 3 ml of chlorophyll
extract to a cuvette and the OD values at 645 nm and
663 nm were read in ELICO UV spectrophotometer
against a DMSO blank. The chlorophyll content in
different treatments was calculated by using the
following formulae,
DNA isolation
DNA extraction protocol followed was according to
Sambrook et al [11], with some modifications. Bacterial
isolates were grown in Lurea broth and incubated at
33oC for overnight under shaking. About 1.5 ml of
culture was taken in micro centrifuge tube, spin for 7
minutes and supernatant was decanted. To the pellet
567µl of TE Buffer, 3µl of 20 mg/ml proteinase-k, 30µl
of 10 per cent SDS were added and incubated for one
hour at 37°C. Again 100µl of 5 M NaCl and 80 µl of
CTAB solution were added and incubated for ten
minute at 65°C. Further it was extracted with equal
volume of Chloroform:Isoamyl alcohol and the aqueous
phase was transferred to the fresh tube and to this
equal volume of Phenol:Chloroform:Isoamyl alcohol
was added and subjected to centrifugation at 8,000 rpm
for 5 min at 4°C. It was washed with chloroform:
Isoamyl alcohol until the clear supernatant was
obtained. Then equal volume of chilled propanol was
added, mixed gently and kept at -20 oC overnight for
precipitation of DNA. Later centrifuged at 10,000 rpm
for 20 min at 4oC to pellet the DNA. The pellet was
washed with 70 per cent ethanol and air-dried. The
DNA was dissolved in TE buffer.
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
PCR amplification conditions
For fingerprinting and diversity analysis, PCR
amplification conditions were optimized based on the
protocol outlined as earlier [13], with minor
modifications.
Reagents used in the PCR
Template DNA: 30 ηg/µl
dNTPs: 2 mM
Taq polymerase: 1 U/µl
Primer : 100µM
10 X Taq assay buffer A: 50mM KCl, 1.5 mM MgCl2 10
mM Tris. HCl pH 9.0, Gelatin 0.1 %, 0.5% Triton-X100
and 0.05% NP40.
30 ηg of genomic DNA was used as the template for the
standardization of PCR reactions and the PCR
conditions were optimized to produce the reproducible
and fine fingerprints. PCR reactions were performed in
a final volume of 25 µl containing 30 ηg of template
DNA, 0.75 µl of 2mM dNTPs each, 2.5µl of 10X taq
buffer,0.36µl 1 unit of Taq DNA polymerase, 3 ml of 10
pico mole Primer. Amplifications were achieved in
MWG-Biotech primus thermocycler with the program
consisting initial denaturation of 94 oC for 3 min
followed by 45 cycles each consisting of denaturation at
94 oC for 1 min, primer annealing temperature at 370
for 30 seconds, primer extension at 72 oC for 3 min,
and a final extension of 72 oC for 10 min. These
reactions were repeated to check the reproducibility of
the amplification.
Selection of primers
To choose the RAPD primers that can amplify
informative sequences, primer screening was carried
out using DNA obtained from the Bacillus megaterium
isolates out of the 7 primers screened. From these 7
primers finally 4 primers producing sharp, intense
bands were selected for the RAPD analysis.
Table 2. RAPD primers with sequences chosen for analysis
S. No.
Sequence
Random primer 1
5’-GGT GCG GGA A-3’
Random primer 2
5’-GTA GTC ATA T-3’
Random primer 3
5’GAG AGC CAA C3’
Random primer 4
5’-GAA CGG ACT C-3’
Agarose gel electrophoresis
Agarose gel electrophoresis was performed to resolve
the amplification product using 1.5 per cent agarose in
1X TBE buffer, 0.5µg/ml of ethidium bromide, and
loading buffer (0.25% Bromophenol Blue in 40%
sucrose). 5 µl of the loading dye was added to 25µl of
PCR products and loaded to the agarose gel.
Electrophoresis was carried at 65 V for 4.5 hour. The
gel was visualized under UV light and documented
using Hero Lab Gel Documentation unit.
Analysis of RAPD data
The bands were manually scored ‘1’ for the presence
and ‘0’ for the absence and the binary data were used
176
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
for statistical analysis. The scored band data (presence
or absence) was subjected to cluster analysis using
STATISTICA. The dendrogram was constructed by
Unweighted Paired Group Average method of
clustering using minimum variance algorithm. The
dissimilarity matrix was developed using Squared
Euclidean Distance (SED), which estimated all the pair
wise differences in the amplification product. Only clear
and unambiguous bands were taken in to account and
the bands were not scored if they were faint or diffused,
as such fragments posses poor reproducibility. The
band sizes were determined by comparing with the 500
bp DNA ladder, which was run along with the amplified
products. Dendrogram was computed based on Ward’s
method of clustering, using minimum variance
algorithm [14].
Principal component analysis (PCA)
This technique help in converting a set of variables in to
a few dimensions using which the genotypes /clones
under study can be depicted in a two or three
dimensional space [15]. Thus, the variations of several
variables will be condensed in to a set of limited axes.
Such a graphical analysis help in identifying the
individuals that tends to cluster together. The genetic
relationships between the different genotypes were
estimated with the PCA developed from dissimilarity
matrix. The genotypes were clustered on the first three
axes and the pattern of clustering or separation of
genotypes from one another was also analyzed.
RESULTS AND DISCUSSION
Of the various rhizosphere associated bacteria, Bacillus
megaterium, is probably the most studied and appear
to have significant potential for commercial application
[16]. Investigations were carried out on Biological and
molecular characterization of Bacillus megaterium
isolated from different rhizosphere soil of Chick pea
plants. The isolates were examined for the genetic
diversity and the biocontrol activity of these isolates on
root pathogenic fungi Pthyium sp. The results obtained
on this investigation are presented below.
Isolation and identification of Bacillus megaterium
The bacterial cultures were isolated from ten agro
climatic zones of Karnataka. The cell material was
checked microscopically for the presence of typical
cells of Bacillus megaterium and purified on nutrient
agar plates. Then all the isolates were subjected to
various tests for confirming their identity.
variability in the solubility zones. Zone 7 showed very
good solubilising ability in Sperber’s media. (plate 2
and table 2).
Screening of Bacillus megaterium isolates on
aerobic rice
Response of Aerobic rice to inoculation of Bacillus
megaterium isolates
Plant Height
The plant height was found to increase steadily with
number of days after inoculation. The height of the
inoculated plants remained always greater than the
uninoculated plants. However the heights differed
significantly among the plants inoculated with various
isolates. The least plant height was recorded in
uninoculated control plants.
Isolation and Identification
Isolation
Isolation of Bacillus megaterium was made from soils of
different agro climatic zones by the enrichment culture
technique in king’s ‘B’ agar medium. All the isolates
were subjected to various tests to confirm identity.
Identification
Colony morphology
Morphologically, all the bacterial isolates exhibited
typical Bacillus megaterium characters. They produced
smooth, bluish green colonies. Most of the isolates
formed well developed colonies on NA medium within
four days of incubation.
Microscopic observation
Bacillus megaterium isolates were further examined for
their Gram’s reaction and shape .Characteristically all
the isolates were gram negative and rod shape.
Figure 1. Colony morphology of Bacillus megaterium
Identification of Bacillus megaterium
Physiological tests
All the physiological tests that were mentioned were
tested and results are presented in the table.
Phosphate solubilising efficiency of different isolates of
Bacillius megaterium
The phosphate solubilising efficiency of different
isolates of Bacillius megaterium was tested on modified
Sperber’s medium. All the isolates found to have good
solubilising ability. However all the isolates showed
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
177
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
Figure 2. Microscopic observation of Bacillus megaterium
Table 2. Biochemical and physiological characters of Bacillus megaterium
S. No.
ISOLATES
1
2
3
4
5
6
7
8
9
10
GL-164
GL-164
GL-265
GL-265
GL-277
GL-277
DDGE-50
DDGE-50
CB-23
CB-23
Growth at
41oC
+
+
+
+
+
+
+
+
+
+
Growth at
4 oC
-
Gelatin
liquefaction
+
+
+
+
+
+
+
+
+
+
Levan
formation
+
+
+
+
+
+
+
+
+
+
Indole
production
-
Biochemical and physiological characters
Gelatin liquefaction
All isolates were found positive to gelatin liquefaction
as indicated by the production of yellowish or bluish
green fluid on the surface of gelatin agar medium.
Figure 5. Levan Formation
Figure 3. Gelation Liquefaction Test
Figure 6. Indole Production
Figure 4. Gelain solidification test
Catalase Test
All isolates were found positive to catalase as indicated
by formation of air bubbles on addition of hydrogen
peroxide.
Levan formation
Test for Levan formation on king’s B agar medium with
3% sucrose had indicated that all isolates produced
slimy colonies due to Levan formation.
Indole production test
In this test all the isolates were found negative as they
did not form a red layer at the top of tryptophan broth
medium.
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
Figure 7. Catalase Test
178
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
10
9
8
7
6
5
4
3
2
1
Figure 8. Plant isolate after 15 days
Growth studies at 4oC and 41oC
At 4oC these isolates do not show growth but show
growth at 410C in terms of turbidity. Based on these
results they are identified as Bacillus megaterium.
Effect of Bacillus megaterium on the growth of
agriculturally important plant (Vigna umbellate)
Bacillus megaterium from ten different agro climate
zones were inoculated to the seeds of Vigna umbellata
in the ratio 1:4 (5ml culture: 20ml water). These plants
were examined for their performance under
greenhouse condition for 45 day at 15 days i.e. 15th,
30th and 45th. The parameters noted were height of
plant, number of leaves and branches and biomass of
plant.
Table 3. Effect of Bacillus megaterium on plant height of
Vigna umbellata
INOCULUM
CONTROL
Zone 1
Zone 2
Zone 3
Zone 4
Zone 5
Zone 6
Zone 7
Zone 8
Zone 9
Zone 10
15th DAY
12.6
12.2
12.2
12.6
11.8
12.2
10.8
16.1
11.6
12.6
14.2
Plant height (cm)
30th DAY
29.7
28.7
32.0
34.7
33.7
33.3
33.0
34.7
33.6
32.0
33.4
45th DAY
176.3
171.8
169
227.3
233.3
126
187.3
206.3
196
166.3
184.6
Table 4. Effect of Bacillus megaterium on number of leaves of
Vigna umbellata
INOCULUM
CONTROL
Zone 1
Zone 2
Zone 3
Zone 4
Zone 5
Zone 6
Zone 7
Zone 8
Zone 9
Zone 10
15th DAY
5
5
5
5
5
5
4
5
5
5
5
Number of Leaves
30th DAY
45th DAY
22
21
20
24
19
22
23
21
20
20
21
60
49
71
51
65
73
56
49
57
58
50
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
Table 5. Effect of Bacillus megaterium on number of Branches
of Vigna umbellata
INOCULUM
Number of Branches
30th Day
45th Day
CONTROL
Zone 1
Zone 2
Zone 3
Zone 4
Zone 5
Zone 6
Zone 7
Zone 8
Zone 9
Zone 10
9
8
9
10
8
9
8
8
8
8
8
23
17
19
18
21
17
17
13
19
16
13
Plant Biomass
In all the inoculated treatments, the plant biomass
increased significantly compared to uninoculated plant
at all the growth intervals.
Table 6. Shoot weight and root weight of Vigna umbellata
INOCULUM
CONTROL
Zone 1
Zone 2
Zone 3
Zone 4
Zone 5
Zone 6
Zone 7
Zone 8
Zone 9
Zone 10
Shoot Weight (g)
Wet
Dry
174.3
45.2
170.1
55.1
186.2
50.9
198.9
57.6
172.8
48.7
164.1
53.3
192.9
50.0
168.5
51.9
153.2
42.6
125.3
41.4
162.8
52.0
Root Weight (g)
Wet
Dry
20.2
10.7
16.4
9.8
29.3
10.8
26.1
10.8
29
10.3
22.7
10.4
26.9
10.4
22.3
10.2
17.8
9.7
20.6
9.4
20.3
12.0
Molecular markers
Standardization of protocol for RAPD analysis
Amplification conditions
For fingerprinting and diversity analysis, PCR
amplification conditions were optimized based on the
protocol outlined by [17, 18] with minor modifications.
In order to obtain high amplification rate and
reproducible banding pattern, different duration for
hot start, denaturation, and primer annealing and
primer extension were tried.
179
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
The PCR reaction was evaluated for 30, 40 and 45
cycles using standard buffer as outlined in Material and
Methods. The optimum conditions for each cycle of PCR
were developed for obtaining high amplification levels
The optimum PCR conditions consisted of the following
steps which were repeated for 45 times. Initial strand
separation or hot start at 940C for three minute
followed by, 45 cycles of
i. Denaturation at 940C for one minute.
ii. Primer annealing at 370C for one minute.
iii. Primer extension at 720C for two minute and
iv. Final extension period at 720C for ten minute
Reaction parameters
It is important to optimize the concentration of PCR
mixture, in order to produce informative and
reproducible RAPD fingerprints. Hence different
concentrations and template DNA (10-15ηg, 25-30ηg
and 40-50ηg) were tried with similar amplification
conditions (Table 4). A concentration of 25-30ηg of
template DNA and 2mM of dNTPs per reaction were
found to be optimum for obtaining intense, clear and
reproducible banding pattern in Bacillus megaterium
isolates. In all these cases, 3 µl of 10 pico moles of
primer and 0.36 µl of 1 unit of Taq polymerase per
reaction were used. However, fluctuation in the
concentration of template DNA did affect the
amplification, with too little DNA (10-15ηg) causing
either reduced or no amplification of small fragments
and higher concentration of DNA (40-50 ηg) producing
a smear.
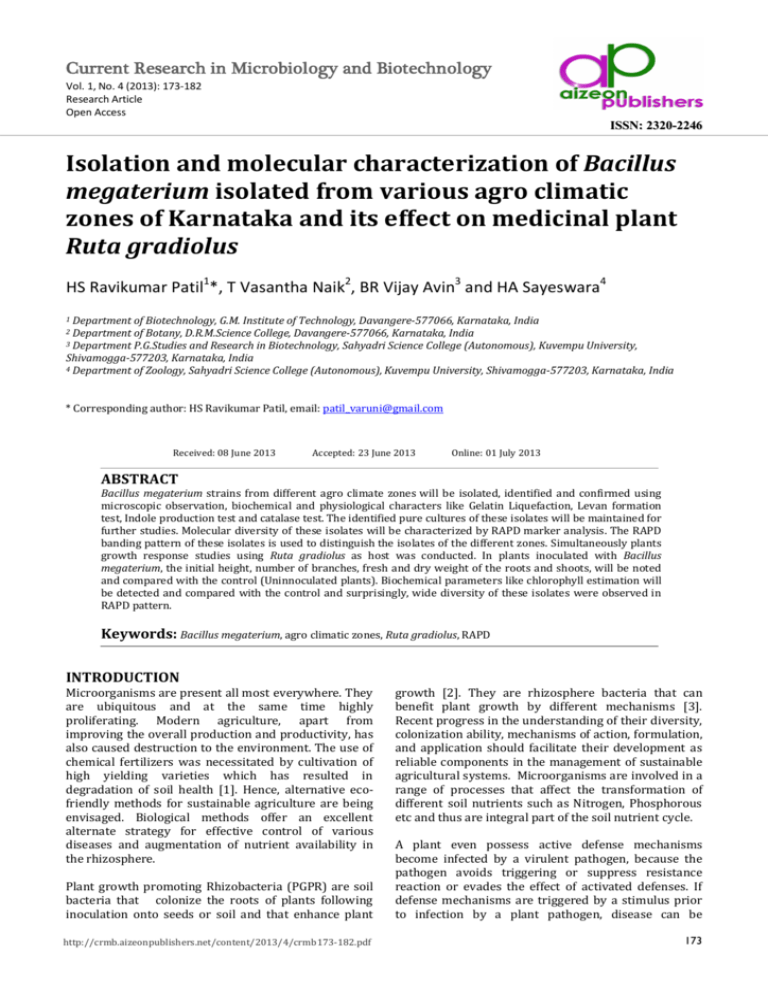
Figure 9. RAPD GEL profile of Bacillus megaterium isolates
generated using 10-mer random primer no.1.lane 1, 2,
3,4,5,6,7,8,9 and 10 are isolates 1 to 10 respectively.
Figure 10. RAPD GEL profile of Bacillus megaterium isolates
generated using 10-mer random primer no.2.lane 1, 2,
3,4,5,6,7,8,9 and 10 are isolates 1 to 10 respectively.
Table 7. Optimum concentration and conditions for RAPD
analysis
Conditions/ concentration
Evaluated
Optimum
Variable
PCR amplification
Hot start
(94 0C)
Denaturation
(94 0C)
Annealing
(40 0C)
Extension
(72 0C)
Number of cycles
RAPD Protocol
Template DNA
dNTPs
2min, 3min, 4 min.
3min,
30sec, 1min, 1.5min.
1min,
1min, 1.5min, 2.0min
1min,
1min, 1.5min, 2.0min,
3min
35, 40, 45 cycles
2min,
10-15ng, 25-30ng, 4050ng
1.5mM, 2mM,
25-30ng
45 cycles
2mM
Table 8. Oligonucleotide primers that showed genetic
variation among the Bacillus megaterium isolates
Primers
Random
primer 1
Random
primer 2
Random
primer 3
Random
primer 4
Total
Percent
Figure 11. RAPD GEL profile of Bacillus megaterium isolates
generated using 10-mer random primer no.3.lane 1, 2,
3,4,5,6,7,8,9 and 10 are isolates 1 to 10 respectively.
No.
of
amplified
fragments
No. of polymorphic
bands
shared
unique
12
07
04
No.
of
Mono
morphic
bands
01
08
04
02
02
10
06
04
00
10
10
00
00
40
27
67.50%
10
25%
03
7.50%
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
Figure 12. RAPD GEL profile of Bacillus megaterium isolates
generated using 10-mer random primer no.4.lane 1, 2,
3,4,5,6,7,8,9 and 10 are isolates 1 to 10 respectively.
CONCLUSION
Bacillus megaterium have emerged as the largest and
potentially most promising group of the plant growth
promoting rhizobacteria (PGPR) with their simple
nutritional requirements, flexible metabolism, their
short generation time and motility. They are well
equipped as primary colonizers of roots. Through
several mechanisms, they promote plant growth and
inhibit soil borne or seed borne phytopathogens. These
include the production of extra cellular growth180
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
promoting
substances
[19],
iron-chelating
siderophores [20], antibiotics [21], HCN [22] and
competition or energy yielding nutrients and space.
Due to their wide spectra of antagonism and
siderophore production, Bacillus megaterium is good
candidates for biological control of phytopathogens,
especially in alkaline soils where Fe availability is
limited. Therefore, a study was undertaken to find the
molecular and physiological variability in Bacillus
megaterium strains isolated from different agro
climatic zones of Karnataka.
Tree Diagram for 10 Variables
Ward`s method
Squared Euclidean distances
GL_164_1
GL_164_2
GL_265_1
GL_277_1
GL_277_2
DDGE_50_
CB_23_1
GL_265_2
DDGE_5_2
CB_23_2
3
4
5
6
7
8
9
10
11
12
Linkage Distance
Figure 13. Dendrogram based on RAPD profile of 10 Bacillus megaterium isolated from different rhizosphere soil of Chick pea
Keeping in view, the Bacillus megaterium strains from
different agro climatic zones were isolated, identified
and confirmed using standard synaptic keys. These
included microscopic observation, biochemical and
physiological characters like Gelatin liquefaction test,
Levan formation test, Indole production test and
Growth studies at 40C and 410C. isolates were identified
and characterized as Bacillus megaterium. Pure cultures
of these isolates were maintained for further studies.
Molecular diversity of these isolates was characterized
by RAPD marker analysis. In the RAPD marker analysis
clearly depicted that all the ten Bacillus megaterium
isolates formed two major clusters. Among the two
major groups, isolate 4, 5, and 6 formed the first group
and the isolate 1, 2, 3, 7, 8, 9 and 10 formed the second
group. The RAPD banding pattern of these isolates
could easily distinguish the isolates of different zones.
Simultaneously, the bio control activity of Bacillus
megaterium was conducted against root pathogenic
fungi Pthyium. Ten different isolates shown the
remarkable growth inhibition of Pthyium fungi.
growth response were grouped together when
compared to the other isolates; these also
belonged to the same group in the tree diagram.
REFERENCES
1.
2.
3.
4.
5.
6.
7.
From the study, the following conclusions were drawn.
Different rhizosphere isolates of Bacillus
megaterium shown bio control activity against
Pthyium.
RAPD banding pattern also cannot be correlated
but the PCA of the RAPD analysis indicated that
the 5 and 6 isolates which gave higher plant
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
8.
Cook, K., (1990), Microbial inoculation in agriculture. Shell
Agriculture, 9: 22 – 25.
Kloepper, J.W., (1983), Effect of seed piece inoculation with
plant growth promoting rhizobacteria on population of
Erwina carotovora on potato roots and in daughter tubers
.Phytopathology, 73: 217-219.
Ganesan, D. Gnanamanickam, S.S., (1987), Biological control
of scleritium rolfsii sacc. In peanut by inoculation with
Pseudomonas fluorescens. Soil biology and Biochemistry, 19:
35 – 38.
Gultterson, N.I., Layton, T.J., Ziegle et al., (1986), Molecular
cloning of genetic determinants for inhibition of fungal
growth by a fluorescent pseudomonad. Journal of
Bacteriological, 165: 696-703.
Mew, T.W and Rosales, A.M., (1986), Bacterization of rice
plants for control of sheath blight caused by Rhizoctonia
Solani. Phytopathology, 76:1260-1264.
Muthukumaraswamy, R., Revathi, G., Vadivelu, M., (2002),
Antagonistic potential of N2-fixing Acetobacter diazotrophicus
against colletotrichum falcahem, A causal organism of red rot
of sugar cane. Curr. Sci., 81(9): 1063-1065.
Bagnasco.P, De La Fuente.L, Gualtieri.G, et al., (1998),
Fluorescent Pseudomonas spp., as bio control agents against
forage legume root pathogenic fungi. Soil Biol.Bioche. 30
(10/11): 1317-1322.
Howell, C.R. and Stipanvic, R.D., (1978), Control of
Rhizoctonia solani on cotton seedlings with Pseudomonas
fluorescens and with an antibiotic produced by the bacterium.
Phytopathology. 64: 480 – 482.
181
HSR Patil et al. / Curr Res Microbiol Biotechnol. 2013, 1(4): 173-182
9.
10.
11.
12.
13.
14.
15.
16.
17.
18.
Buchanan, R.E. And Gibbons, N.E., (1974), Bergey’s manual of
determinative bacteriology, 8th Edn, Williams and Wilkins,
Baltimore, pp-217
Bension, H.J., (1990), Microbiological Applications fifth
edition, Wm.C.Brown Publishers, London, pp.155
Sambrook, J., Fristch, E.F. and Maniatis, T., (1989), Molecular
cloning: A laboratory manual 2nd edition CSH laboratory press
Vol. I, Section 6.
Harrison, L.A., Letrendre, I., Kovacevich, P, et al., (1993), A
comparison of methods for measuring the colonization of
root system by fluorescent Pseudomonas. Soil Biology and
Biochemistry, 25: 215
Burr, T.J., Schroth, M.N. and Suslow, T., (1978), Increased
potato yields by treatment of seed pieces with specific strains
of Pseudomonas fluorescens and P. putida. Phytopathology, 68:
1377 – 1383.
Weller, D.M., (1988), Biological control of soil borne plant
pathogens in the rhizosphere with bacteria. Annual Review of
Phytopathology, 26: 379 – 407.
Kloepper, J.W., R. Lifshiftz AND R.M. Zablotowicz, (1989), Free
living bacterial inocula for enhancing crop productivity.
Trends in biotech.7: 39-44.
Baumforth, K.R.N., Nelson, P.N., Digby, J.E., et al., (1999),
Polymerase chain reaction. Mol. Pathol., 52: 1-10.
Williams, J.G., Kubelic, A.R., Livak, KL, et al., (1990), DNA
polymorphism amplified by arbitrary primers are useful as
genetic markers. Nucl. Acids, Res., 18: 6513.
Welsh, J. and MC Clelland, M., (1990), Fingerprinting genomes
using PCR with arbitrary primers. Nucl. Acids. Res., 18 (24):
7213-7218
19. YUEN, G.Y. and Schroth, M.N., (1986), Interaction of
Pseudomonas fluorescens strain E6 with ornamental plants
and its Effects on the composition of root colonizing
microflora .phytopathology , 76 : 176 -180.
20. Johri, B.N., Alok Sharma (2006), In: Plant growth-promoting
Pseudomonas sp. Strains reduce natural occurrence of
Anthracnose in Soybean (Glycine max L.) in Central
Himalayan Regin, 52: 390-394.
21. David C.Sands and Lester Hankin (1975), Ecology and
Physiology
of
fluorescent
pectolytic
Pseudomonas,
Phytopathology, 65:
22. Weller, D.M., Howke, W.J. and Cook, R.J., (1988), Biological
control of soil borne plant pathogens in the rhizosphere with
bacteria Phytopathology, 38:1094.
23. Voisard, C., Keel, O. Haas. and G. Defago., (1989), Cyanide
production by Pseudomonas fluorescens helps to suppress
black root rot of tobacco under gnotobiotic condition
European Microbiological Journal, 8: 351 – 358
© 2013; AIZEON Publishers; All Rights Reserved
This is an Open Access article distributed under the terms of
the Creative Commons Attribution License which permits
unrestricted use, distribution, and reproduction in any medium,
provided the original work is properly cited.
*****
http://crmb.aizeonpublishers.net/content/2013/4/crmb173-182.pdf
182