Enzyme Lab
advertisement

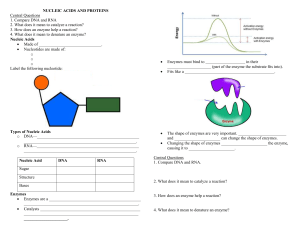
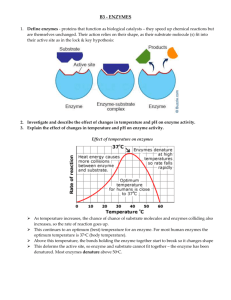
MA 61 Precalculus for Biology Majors Lab II: Creating, Identifying, and Interpreting Graphs of Data A primary goal of biologists is to find quantifiable patterns in nature so that they can understand and make predictions regarding the underlying phenomena and processes they are studying. An important set of skills to develop, is creating, identifying, and interpreting graphs of data. In this lab we will learn how to create, identify and interpret graphs of functions using Excel. We need to become familiar with various families of functions, their associated graphs and transformations on these functions to obtain new functions. 1. Polynomial Functions: Linear (1st degree): nd Quadratic (2 degree): Cubic (3rd degree): Quartic (4th degree): Quintic (5th degree): f (x) mx b f (x) ax 2 bx c f (x) ax 3 bx 2 cx d f (x) ax 4 bx 3 cx 2 dx e f (x) ax 5 bx 4 cx 3 dx 2 ex f f (x) a n x n a n 1 x n 1 a n 2 x n 2 nth Degree: + a 2 x 2 a1x a 0 also written as pn (x) Even: Odd: Only even powers of x and constants Only odd powers of x and no constant term 2. Exponential Functions: Increasing: f (x) a er x , f (x) a cr x Decreasing: f (x) a e r x , f (x) a c r x p (x) a a ax , f (x) , f (x) , in general f (x) n x bx c bx c pm (x) 3. Rational Functions: f (x) 4. Power Functions: f (x) a x p q , or in radical form f (x) a p x q (root functions) 5. Logarithmic Functions: Will discuss these later in the course 6. TrigonometricFunctions: Will discuss these later in the course Common Transformations: 1. Vertical Shift: 2. Horizontal Shift: 3. Vertical Stretch: 4. Vertical Compression: 5. Horizontal Stretch: f (x) f (x) c , if c>0 it moves up if c<0 it moves down f (x) f (x c) , if c>0 it moves right, if c<0 it moves left f (x) a f (x) , a>1 f (x) a f (x) , 0<a<1 f (x) f (ax) , 0<a<1 1 6. Horizontal Compression: 7. Reflection about the x-axis: 8. Reflection about the y-axis: f (x) f (ax) , a>1 f (x) f (x) f (x) f ( x) Lab Assignment Part 1: Working with Functions: Their Graphs and Transformations A. Use the Excel spreadsheet provided (MultiGraph.xls) to obtain graphs of the following: 1. y1 f (x) x 3 y2 f (x) 2 x 1 y3 f (x) 0.5 x 1 Describe what happened to the linear function as we changed m and b, f (x) m x b . What are the Domains and Ranges of the functions? 2. y1 f (x) x 2 y2 f (x) x 2 3x 10 y3 f (x) x 2 3x 10 Describe what happened to the base function, f (x) x 2 . What are the Domains and Ranges of the functions? 3. y1 f (x) x 3 y2 f (x) 3x 3 6x 2 15x 18 y3 f (x) -3x 3 6x 2 15x 18 Describe what happened to the base function, f (x) x 3 . What are the Domains and Ranges of the functions? 4. y1 f (x) e x y2 f (x) e2 x y3 f (x) e0.5 x Describe what happened to the base function, f (x) e x . What are the Domains and Ranges of the functions? 5. y1 f (x) e x y2 f (x) 2 e x y3 f (x) 0.5 e x Describe what happened to the base function, f (x) e x . What are the Domains and Ranges of the functions? 6. y1 f (x) e x y2 f (x) e x y3 f (x) e2 x Describe what happened to the base function, f (x) e x . 2 What are the Domains and Ranges of the functions? 7. y1 f (x) e x y2 f (x) -e x y3 f (x) -2 e x Describe what happened to the base function, f (x) e x . What are the Domains and Ranges of the functions? B. Use the Excel spreadsheet provided (MultiGraph.xls) to obtain graphs of the following transformations of the base functions: 1. y1 f (x) x 2 y2 f (x) x 2 2 y3 f (x) x 2 2 Describe what happened to the base function, f (x) x 2 . 2. y1 f (x) x 2 y2 f (x) (x 2) 2 y3 f (x) (x 2) 2 Describe what happened to the base function, f (x) x 2 . 3. y1 f (x) x 2 y2 f (x) 2 x 2 y3 f (x) 0.5 x 2 Describe what happened to the base function, f (x) x 2 . 4. y1 f (x) x 2 y2 f (x) (2x)2 y3 f (x) (0.5 x) 2 Describe what happened to the base function, f (x) x 2 . 5. y1 f (x) x 2 y2 f (x) x 2 y3 f (x) ( x)2 Describe what happened to the base function, f (x) x 2 . 6. y1 f (x) x 3 y2 f (x) x 3 y3 f (x) ( x)3 Describe what happened to the base function, f (x) x 3 . 3 Lab Assignment Part 2: Rational Functions in Biology Basics of Enzyme Kinetics Enzymes are organic catalysts that control the rate of chemical reactions in cells. In general, enzymes speed up the rate of reaction by lowering the activation energy required to start reactions. Enzymes are extremely efficient. They can catalyze reactions at rates up to 10 billion times higher than comparable non-catalyzed reactions. The chemistry in biological organisms, however, is fundamentally different from that in test tubes and in chemical engineering largely due to the unique roles played by enzymes. Living systems depend on chemical reactions, which on their own, would occur at extremely slow rates. Enzymes are catalysts, which reduce the needed activation energy so these reactions proceed at rates that are useful to the cell. With the presence of enzymes as specific catalysts, biochemical reactions are greatly accelerated. The unique role of enzymes also makes the kinetic of such reactions significantly different from that of conventional chemical kinetics. The key difference between the enzyme kinetics and the conventional bimolecular reactions is the large excess of substrate concentration with respect to the enzyme concentration In typical enzyme-catalyzed reactions, reactant and product concentrations are usually hundreds or thousands of times greater than the enzyme concentration. Consequently, each enzyme molecule catalyzes the conversion to product, of many reactant molecules. The kinetics of simple reactions in the body, were first characterized by biochemists Michaelis and Menten. The concepts underlying their analysis of enzyme kinetics continue to provide the cornerstone for understanding metabolism today, and for the development and clinical use of drugs aimed at selectively altering rate constants and interfering with the progress of disease states. The Michaelis-Menten equation, V VMax S S K m is a quantitative description of the relationship among the rate of an enzyme – catalyzed reaction [V], the concentration of substrate [S] and two constants, VMax and Km (which are constant for the particular equation). The symbols used in the Michaelis-Menton equation refer to the reaction rate [V], maximum reaction rate (VMax), substrate concentration [S] and the Michaelis-Menton constant (Km). Now this is not the most useful form of the equation. We often define v V / Vmax , to make things easier to understand and work with. This is called scaling a variable. Then the Michaelis-Menten equation becomes: S v S K m The Michaelis-Menten equation can also be used to demonstrate that at the substrate concentration that produces exactly half of the maximum reaction rate, i.e.,1/2 VMax, the substrate concentration is numerically equal to Km. This fact provides a simple yet powerful bioanalytical tool that has been used to characterize both normal and altered enzymes, such as those that produce the symptoms of genetic diseases. Furthermore, the activity of enzymes is strongly affected by changes in pH and temperature. 4 Each enzyme works best at a certain pH and temperature (see graphs below), its activity decreasing at values above and below that point. A. Use the Excel spreadsheet provided (MichaelisMentenLab.xls) to obtain graphs of the scaled Michaelis-Menten Equation: S v S K m Note: v will range from 0 to 1. a value of 1 corresponds to the maximum reaction rate, Vmax. Let S range from 0 to 50 mM Vary Km as: 1, 2, 4, and 8mM Answer the following: 1. Describe what happens to v as you change Km. 2. For what value of S is v equal to half the maximum reaction rate? 3. What happens as S ? B. We can often combine the various base functions listed above to make new functions. For example, say we wished to find functions that give them same curves illustrated for the reaction rate dependence on both pH and temperature given above. m x n x For temperature dependence (graph) shown above consider the function: f (x) a e b , where b x represents temperature in degrees Celsius and f(x) is the reaction rate. 1. Use the Excel spreadsheet provided (MultiGraph.xls) to graph this function for a = 5, b = 60, m = 0.6 and n = 12. Let x range from 0 to 80. 2. For what temperature (x value) is the reaction rate the largest? 3. Why did we choose these values for x (recall the physical meaning of x)? 4. Compare the results to the graph given above. 5. Vary a, b, m, and n to see how the graph changes. C. For the pH dependence (graph) shown above consider the function: f (x) a e pH and f(x) is the reaction rate. x b c 2 , where x is the 1. Use the Excel spreadsheet provided (MultiGraph.xls) to graph this function for a = 4, b = 8, and c = 3. Let x range from 0 to 14. 2. For what pH (x value) is the reaction rate the largest? 3. Why did we choose these values for x (recall the physical meaning of x)? 4. Compare the results to the graph given above. 5. Vary a, b m and n to see how the graph changes. 5