1747-1028-4-20-S2
advertisement

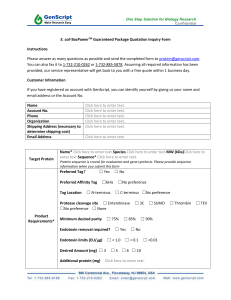
Additional File 2, Comparison of different proteomics-based techniques METHOD DESCRIPTION ADVANTAGES DISADVANTAGES SENSITIVITY 2D GEL ELECTROPHORESIS/MASS SPECTROMETRY Separation of Ability to identify Proteins expressed at Detection complex proteins unknown proteins low abundance may be sensitivity is in the via 2D gel Detects protein missed nanogram range electrophoresis modification Unsuited for (50 ng/spot for based charge and (phosphorylation diagnostic application Coomassie Blue; size and methylation) Limited 1 ng/spot for silver Major protein Used for various reproducibility and stain) identification by MS biological samples, high rate of false Using fluorescent Detects about 2000including tissue, identification 2D-differential gel 2500 spots/gel blood and other Limited dynamic electrophoresis biological fluids range (2D-DIGE), semi-quantitative sensitivity improves by 10 fold (CyDye label) LIQUID CHROMATOGRAPHY/MASS SPECTROMETRY LC to separate Ability to identify Proteins expressed at Detection proteins in a sample, unknown proteins low abundance may be sensitivity is in the with sequential LC improved missed nanogram range for improved separation Unsuited for or ~20 cells separation efficiency efficiency diagnostic application 1% false positive MS to compared to 2D gel Limited rate systematically Used for various reproducibility and identify the major biological samples, high rate of false proteins including tissue, identification Detects over 1000 blood and other Limited dynamic proteins/run biological fluids range semi-quantitative PROTEIN ARRAY Individual protein immobilization on a solid-support (glass or membrane) Individual proteins identified by labeled antibodies Detects over 1000 proteins/array Multiple whole-cell or tissue lysate immobilization on individual spots on a solid support (similar to tissue microarray format) Presence of specific proteins are detected by antibody Detects < 100 proteins/array High sensitivity and Limited protein specificity availability from Good quantitation complex protein range production process High (expression and throughput/density purification) amenable for Limited access to a automation large number of Economical and affinity antibodies for low sample detection. consumption Lots of data from single experiment Software and hardware tools may be shared with DNA microarray REVERSE PHASE PROTEIN ARRAY Highly sensitive Detection sensitivity detection of may be compromised proteins from loss native High throughput, protein conformation i.e. a large number when surface spotted of samples on one Limited sensitivity to slide detect low abundance Minimal sample proteins required Specificity may be Reduced number of compromised from antibodies needed non-specific antibody to detect protein binding (i.e. potential for high background) Limited number of available signaling protein-specific antibodies ANTIBODY ARRAY Detection sensitivity is in the ng/ml range Detection sensitivity is in the picogram range Increased sensitivity Using laser capture microdissection, lysates can be analyzed with as few as 10 cells Capture antibodies are spotted and fixed on a solid surface Proteins (antigens) are captured on the array surface and detected by a second antibody specific for a different epitopes than capture antibody (sandwich format) Detects < 100 proteins/array Complex proteins in a sample (cells or tissue) are separated via gel electrophoresis Proteins then transfers to nitrocellular membrane Proteins detected by multichannel immunoblot (similar to Western Blot) Detects up to 300 proteins/run Highly specific Protein complexity Detection from dual antibody and denaturation may sensitivity is in the detection affect antigenlow pg/ml range Highly sensitive antibody interaction High throughput Need for high-affinity and amenable for and specific antibodies automation for capture and Possible to detect detection protein Limited dynamic modifications range of 2 or 3 orders (phosphorylation, of magnitude methylation, etc) by modificationspecific antibodies Suitable for clinical applications PATHWAY ARRAY Highly sensitive Limited availability of Detection limit of with detection of signaling-related 1 ng for each low abundance antibodies protein with proteins Relative low throughchemiluminescenc Highly specific (as put (one sample per e; 0.1 ng with determined by gel fluorescence immunoreactivity Limited dynamic Linear detection and size) range of 2 or 3 orders range is 100 fold High accuracy and of magnitude for ECL and 1,000 reproducibility for fluorescence. Minimal antibody required for each sample Detects protein modifications (phosphorylation, methylation, etc) BEAD-BASED ARRAY Either capture Highly sensitive Protein complexity Detection limit is antibody or proteins and specific and denaturation sufficient to are coated on beads High throughput affecting antigencapture low Detection of and amenable for antibody interaction abundance protein proteins by labeled automation Need for high-affinity analytes down to antibodies (similar Detects protein and specific antibodies the pg/mL range to antibody array or modifications for capture and ELISA) (phosphorylation, detection Detects 50-100 methylation, etc) by Limited dynamic proteins/run modification range of 2 or 3 orders specific antibodies of magnitude Suitable for clinical applications METHOD DESCRIPTION ADVANTAGES DISADVANTAGES SENSITIVITY 2D GEL ELECTROPHORESIS/MASS SPECTROMETRY Separation of Ability to identify Proteins expressed at Detection complex proteins unknown proteins low abundance may be sensitivity is in the via 2D gel Detects protein missed nanogram range electrophoresis modification Unsuited for (50 ng/spot for based charge and (phosphorylation diagnostic application Coomassie Blue; size and methylation) Limited 1 ng/spot for silver Major protein Used for various reproducibility and stain) identification by MS biological samples, high rate of false Using fluorescent Detects about 2000including tissue, identification 2D-differential gel 2500 spots/gel blood and other Limited dynamic electrophoresis biological fluids range (2D-DIGE), semi-quantitative sensitivity improves by 10 fold (CyDye label) LIQUID CHROMATOGRAPHY/MASS SPECTROMETRY LC to separate Ability to identify Proteins expressed at Detection proteins in a sample, unknown proteins low abundance may be sensitivity is in the with sequential LC improved missed nanogram range for improved separation Unsuited for or ~20 cells separation efficiency efficiency diagnostic application 1% false positive MS to compared to 2D gel Limited rate systematically Used for various reproducibility and identify the major biological samples, high rate of false proteins including tissue, identification Detects over 1000 blood and other Limited dynamic proteins/run biological fluids range semi-quantitative PROTEIN ARRAY Individual protein immobilization on a solid-support (glass or membrane) Individual proteins identified by labeled antibodies Detects over 1000 proteins/array Multiple whole-cell or tissue lysate immobilization on individual spots on a solid support (similar to tissue microarray format) Presence of specific proteins are detected by antibody Detects < 100 proteins/array High sensitivity and Limited protein specificity availability from Good quantitation complex protein range production process High (expression and throughput/density purification) amenable for Limited access to a automation large number of Economical and affinity antibodies for low sample detection. consumption Lots of data from single experiment Software and hardware tools may be shared with DNA microarray REVERSE PHASE PROTEIN ARRAY Highly sensitive Detection sensitivity detection of may be compromised proteins from loss native High throughput, protein conformation i.e. a large number when surface spotted of samples on one Limited sensitivity to slide detect low abundance Minimal sample proteins required Specificity may be Reduced number of compromised from antibodies needed non-specific antibody to detect protein binding (i.e. potential for high background) Limited number of available signaling protein-specific antibodies ANTIBODY ARRAY Detection sensitivity is in the ng/ml range Detection sensitivity is in the picogram range Increased sensitivity Using laser capture microdissection, lysates can be analyzed with as few as 10 cells Capture antibodies are spotted and fixed on a solid surface Proteins (antigens) are captured on the array surface and detected by a second antibody specific for a different epitopes than capture antibody (sandwich format) Detects < 100 proteins/array Complex proteins in a sample (cells or tissue) are separated via gel electrophoresis Proteins then transfers to nitrocellular membrane Proteins detected by multichannel immunoblot (similar to Western Blot) Detects up to 300 proteins/run Highly specific Protein complexity Detection from dual antibody and denaturation may sensitivity is in the detection affect antigenlow pg/ml range Highly sensitive antibody interaction High throughput Need for high-affinity and amenable for and specific antibodies automation for capture and Possible to detect detection protein Limited dynamic modifications ranges of 2 or 3 orders (phosphorylation, of magnitude methylation, etc) by modificationspecific antibodies Suitable for clinical applications PATHWAY ARRAY Highly sensitive Limited availability of Detection limit of with detection of signaling-related 1 ng for each low abundance antibodies protein with proteins Relative low throughchemiluminescenc Highly specific (as put (one sample per e; 0.1 ng with determined by gel fluorescence immunoreactivity Limited dynamic Linear detection and size) ranges of 2 or 3 orders range is 100 fold High accuracy and of magnitude for ECL and 1,000 reproducibility for fluorescence. Minimal antibody required for each sample Detects protein modifications (phosphorylation, methylation, etc) BEAD-BASED ARRAY Either capture Highly sensitive Protein complexity Detection limit is antibody or proteins and specific and denaturation sufficient to are coated on beads High throughput affecting antigencapture low Detection of and amenable for antibody interaction abundance protein proteins by labeled automation Need for high-affinity analytes down to antibodies (similar Detects protein and specific antibodies the pg/mL range to antibody array or modifications for capture and ELISA) (phosphorylation, detection Detects 50-100 methylation, etc) by Limited dynamic proteins/run modification ranges of 2 or 3 orders specific antibodies of magnitude Suitable for clinical applications