Ocean Data View – Presentations and Exercises
advertisement
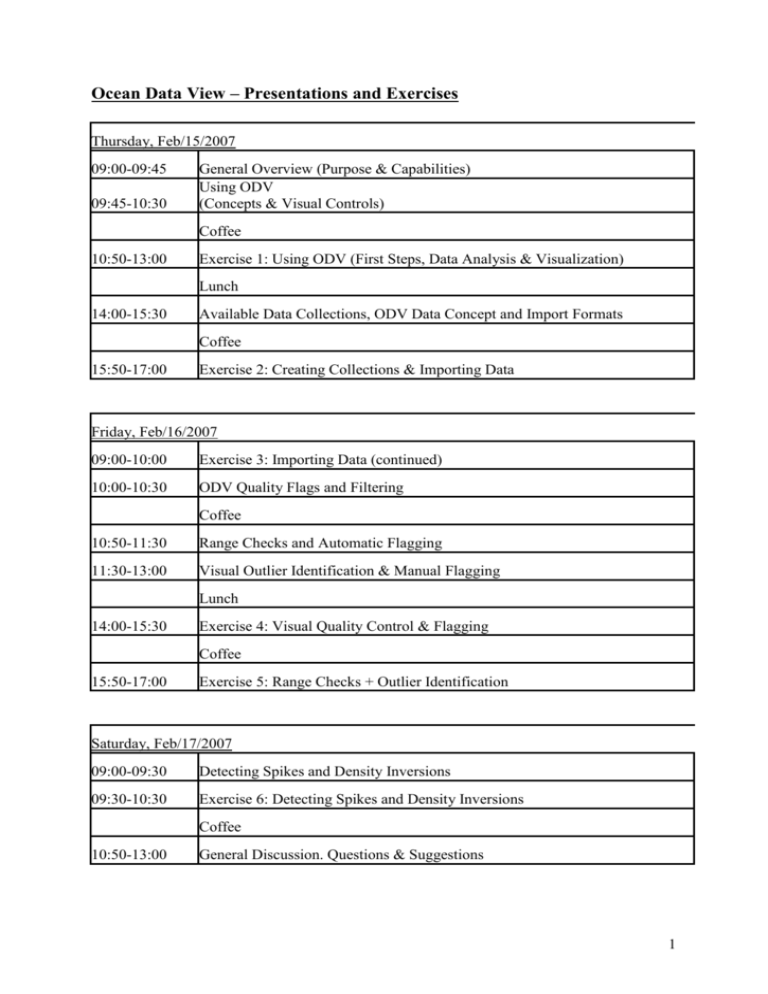
Ocean Data View – Presentations and Exercises Thursday, Feb/15/2007 09:00-09:45 09:45-10:30 General Overview (Purpose & Capabilities) Using ODV (Concepts & Visual Controls) Coffee 10:50-13:00 Exercise 1: Using ODV (First Steps, Data Analysis & Visualization) Lunch 14:00-15:30 Available Data Collections, ODV Data Concept and Import Formats Coffee 15:50-17:00 Exercise 2: Creating Collections & Importing Data Friday, Feb/16/2007 09:00-10:00 Exercise 3: Importing Data (continued) 10:00-10:30 ODV Quality Flags and Filtering Coffee 10:50-11:30 Range Checks and Automatic Flagging 11:30-13:00 Visual Outlier Identification & Manual Flagging Lunch 14:00-15:30 Exercise 4: Visual Quality Control & Flagging Coffee 15:50-17:00 Exercise 5: Range Checks + Outlier Identification Saturday, Feb/17/2007 09:00-09:30 Detecting Spikes and Density Inversions 09:30-10:30 Exercise 6: Detecting Spikes and Density Inversions Coffee 10:50-13:00 General Discussion. Questions & Suggestions 1 Exercise 1: 1. Copy the directories under \SeaDataNet\Reiner Schlitzer to a work directory on your local hard disk. 2. Install the optional package odvmpOP_coastRegional_w32.zip from http://odv.awi.de. This package contains high resolution regional coastlines for the Mediterranean, Baltic, North Sea and other regions. 3. Run ODV and open the MedatlasII_Btl collection on your hard disk. Make the map to use the MeditHR coastlines installed during step 2. Invoke ODV Help and read the chapter on map Display Options for instructions on how to do this. 4. In MAP mode produce full screen maps of (a) all stations in MedatlasII_Btl, (b) of all stations containing data for Phosphate, and (c) of all stations containing data for Phosphate and Nitrate. Save the configuration of (a). Use a descriptive name for the configuration, such as AllStationsMap. How many stations have Phosphate and Nitrate data? 5. Load the AllStationsMap configuration saved during (4) and make Display Option changes, e.g., size and color of station dots, map projection, coastline/bathymetry layers etc. Save your favourite configuration. Undo last configuration changes. 6. Switch to STATION mode and produce plots with temperature, salinity and oxygen profiles. Change the window layout so that you only have two large windows. Change the variables on x and y axis. What are typical oxygen values in 200 m depth in the Black Sea and Meditterranean? 7. (Advanced) Specify a station selection rectangle or polygon to select only stations in a region of interest. Define Potential Temperature as a derived variable. Switch to SCATTER mode and produce a large T/S diagram with salinity on x and Tpot on y. Define Potential Density as second derived variable and define a second window showing profiles of potential density. Do you see any suspicious data? 8. (Advanced) Switch to STATION mode and undo any regional station selection from step 7. Switch to SECTION mode and define your favourite section. Produce section plots of potential temperature, salinity and oxygen. Also create a T/S diagram of the section data. Add contour lines to the section plots and produce GIF, PNG or JPG image output. Exercise 2: 1. Create a new directory profileCollection on your local hard disk and create a new ODV collection in this directory using ODV’s File>New option. Choose a name for the new collection, e.g., MyProfiles. When prompted for the definition of variables in the collection choose Medatlas Bottle variables. Note that the newly created collection is empty, initially. 2. Import some Medatlas bottle data from file b0653311.987 in the \SeaDataNet\Reiner Schlitzer\importData directory. Use option Import>Medatlas Format Files>Profile Data>Single File. Then use option Full Domain from the map’s popup menu to adjust the domain covered by the imported stations. Undo last change before proceeding. 3. (Advanced) Import data from the US NODC World Ocean Database 2005 file OSDO1985.gz. Use option Import>NODC Formats>World Ocean Database>Single File. On the Import Options dialog note that the WOD 2005 import file provides values 2 for Depth [m] while the collection stores Pressure [decibars]. Therefore, depths have to be converted to pressure values during import. Use the ODV help to find out how to do this. Don’t forget to associate or convert other variables as well. Also note that units for the nutrients in the source file and target collection appear to be different. Do we need to convert these? 4. (Advanced) Download ARGO float profile data from http://www.coriolis.eu.org/cdc/argo.htm and import them using Import>ARGO Formats>Float Profiles (…)>Single File. 5. Create a new directory time-seriesCollection on your local hard disk and create a new ODV collection in this directory using ODV’s File>New option. Choose a name for the new collection, e.g., MyTimeSeries. When prompted for the definition of variables in the collection choose Medatlas Time Series variables. Note that the newly created collection is empty, initially. 6. Import some Medatlas current meter data from file example_medatlas_time_series.dat in the \SeaDataNet\Reiner Schlitzer\importData directory. Use option Import>Medatlas Format Files>Time Series Data>Single File. Then use option Full Domain from the map’s popup menu to adjust the domain covered by the imported stations. Undo last change before proceeding. 7. Produce plot of current speed versus time. Exercise 3: 1. If not already present create an ODV desktop icon. 2. Drag the file data_from_WoceBtl.txt from \SeaDataNet\Reiner Schlitzer\importData\speadsheet onto the ODV icon. Note that ODV created a new collection data_from_WoceBtl and imported the data from the .txt file in a single, automatic step. This was possible because data_from_WoceBtl.txt adheres to the ODV Generic Spreadsheet format. Any such file can be simply dragged onto the ODV icon and can be imported automatically. 3. Drag your own data file onto the ODV icon. Is it imported automatically? Does it adhere to the ODV Generic Spreadsheet format? (See help file for format specification.) Ask for help if you are stuck. 3 Exercise 4: 1. Drag the file data_from_WoceBtl_compact_noQF.txt from \SeaDataNet\Reiner Schlitzer\qualityCheck onto the ODV icon. Produce section plots of Nitrate and Silicate with the unchecked data (Display Options settings: Gridded Field, VG Gridding, 30 and 25 for X and Y scale lengths). Save these sections to image files for comparison with the cleaned sections later. 2. Switch to SCATTER mode and visually quality control the data. Note that you can use different kinds of plots to identify outliers, e.g., profiles, T/S plots or general property/property plots, such as for instance Phosphate versus Oxygen or Phosphate versus Nitrate. Some cases are difficult to decide and you may want to look at several plots simultaneously. Start with quality control of Nitrate and Silicate, then do the other variables if time permits. 3. Exclude Questionable and Bad data by applying data quality filters and redo the Nitrate and Silicate sections. Any improvement? 4. Apply visual data quality control to your own dataset. Exercise 5: 1. Drag the file xbt_noQF.txt from \SeaDataNet\Reiner Schlitzer\qualityCheck onto the ODV icon. Switch to SCATTER mode and make sure you have a large plot window with temperature profiles. 2. Note that some XBT locations fall on land. Lets assume you know the correct longitude/latitude values. Edit the metadata of these XBTs accordingly using the Edit Header option of the text window popup menu. 3. Start cleaning the XBT data by applying range checks. 4. By clicking into the data plot try to find profiles that look good over some depth range but have questionable or bad data above or below (if you can’t find one select station # 1248). Do questionable or bad quality flagging for the entire set of problematic values. Note that it may be useful to select a regional subset of stations before doing the quality control. Exercise 6: 1. Open the xbt_noQF collection from exercise 5 again. 2. Define the Vertical Derivative of Temperature as a derived variable and create a window layout with one large plot for temperature profiles and another window with d/dz Temperature versus depth. 3. In the latter window click on points with very large positive or negative d/dz Temperature values to identify spikes. Manually edit the quality flags of the bad data. 4







![A Further Study on Using ˙x = λ [αR +... (P = F − R(F · R)/kRk ]](http://s2.studylib.net/store/data/011582898_1-2f1ec2f71dc156ec0f65b4d813e5066b-300x300.png)
