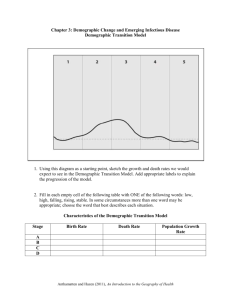
Figure S1 (a) The chloroplast haplotype network of Callitris
advertisement

Figure S1 (a) The chloroplast haplotype network of Callitris columellaris species complex, reconstructed by median joining network method using NETWORK v. 4.6.0.0 [1] with C. endlicheri (GenBank: AB723699) as an outgroup. The size of the circles in the haplotype network is proportional to the haplotype frequency, and missing haplotypes are indicated by rectangles. (b) The neighbour-joining tree of the twelve genetic clusters were estimated by STRUCTURE v. 2.3 [2] based on nuclear EST-SSR genotype data. The circle size is proportional to the expected heterozygosity of each cluster. The colours used in the network and tree match those in figure 2. (a) (b) C. endlicheri 10 steps 0.01 Figure S2 The log-likelihood of the data L(K), and the second-order rate change of L(K) (ΔK) [3] are plotted against the number of genetic clusters (K) assumed in the STRUCTURE analysis [2]. Although ΔK showed a clear peak at K=2, its clustering pattern was too simple to even delineate any morphological species, and not consistent with the structure in the distance-based network in figure 2c. On the other hand, the L(K) increased consistently with increasing number of clusters and reached a plateau around K=10–15. The distribution of the individual trees assigned to given clusters in K=10–15 was geographically structured, which was compatible with both the nuclear genetic structure estimated by the distance-based network and chloroplast DNA haplotypes (figure 2a). Among the plausible number of genetic clusters, we consider K=12 to be the smallest K that can capture most of the structure in the data and which seemed biologically sensible [4], because further clustering of K>12 made little change to the overall genetic structure. Figure S3 Geographic distribution of the summary statistics used in the demographic analysis based on nuclear EST-SSR markers [(a) mean number of alleles across loci (Na), (b) expected heterozygosity (He), (c) mean allele size variance across loci (VAR) and (d) mean M index across loci (MGW)]. The distribution of the species complex is indicated by light-grey colour shading. (a) (b) He Na 0.21 - 0.25 0.25 - 0.29 0.29 - 0.32 1.70 - 2.12 2.12 - 2.53 2.53 - 2.95 0.32 - 0.36 2.95 - 3.37 0.36 - 0.40 3.37 - 3.78 0.40 - 0.44 3.78 - 4.20 (c) (d) MGW ASV 0.80 - 1.68 1.68 - 2.56 2.56 - 3.45 3.45 - 4.33 4.33 - 5.21 5.21 - 6.10 0.54 - 0.62 0.62 - 0.69 0.69 - 0.77 0.77 - 0.85 0.85 - 0.92 0.92 - 1.00 Figure S4 Geographic distribution of the median value of log-transformed ratio of population size (NC / NA) in the demographic analysis based on nuclear EST-SSR markers. The population size change ratio is proportional to the size of circle, and the colour of circles are conditioned by the selected demographic scenarios (light blue: “reduction”, white: “no change” or “indecisive”, and pink: “expansion”). The interior arid zone is indicated by light-grey colour blobs. Population size change ratio 0.2 1.0 3.0 10.0 Figure S5 The results of demographic analyses for pooled population data using an Approximate Bayesian Computation method implemented in DIYABC-v1.0.4.46beta [5]. (a) Population size change ratio for pooled populations (red: mostly arid, blue: monsoon tropics, gray: mostly temperate). (b) Converted time parameters for the populations in which the stable scenario was not rejected. The two broken gray lines represent the time envelopes when generation time was assumed as its minimum (20 years) and maximum (70 years), respectively. (c) Posterior distribution of the effective population size for each population group (black: NC, gray: NA). The broken line shows prior distribution. Population groups which showed significant deviation from “size change scenario” are indicated by asterisks. Figure S6 The results of demographic analyses for pooled population data using a Bayesian method implemented in msvar1.3 [6]. (a) Population size change ratio for pooled populations (red: mostly arid, blue: monsoon tropics, gray: mostly temperate). (b) Converted time parameters for the population. The two broken gray lines represent the time envelopes when generation time was assumed as its minimum (20 years) and maximum (70 years), respectively. (c) Posterior distribution of the effective population size for each population group (black: NC, gray: NA). The broken line shows prior distribution. Note that MCMC chains did not converge for one population group [Ccol (EC)], as indicated by asterisk. Figure S7 Population size change ratio of the ten replicated data sets simulated under a demographic scenario of 10× population expansion. The black and gray lines respectively represent the density estimates using an Approximate Bayesian Computation method implemented in DIYABC-v1.0.4.46beta [5] and a Bayesian method implemented in msvar1.3 [6]. The vertical broken line shows the expected change ratio [log10(10) = 1.0]. References 1. Bandelt H.J., Forster P., Rohl A. 1999 Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16(1), 37-48. 2. Pritchard J.K., Stephens M., Donnelly P. 2000 Inference of population structure using multilocus genotype data. Genetics 155(2), 945-959. 3. Evanno G., Regnaut S., Goudet J. 2005 Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8), 2611-2620. 4. Pritchard J.K., Wen X., Falush D. 2009 Documentation for structure software: Version 2.3. http://pritchbsduchicagoedu/structure_software/release_versions/v232/structure_docpdf. 5. Cornuet J.M., Santos F., Beaumont M.A., Robert C.P., Marin J.M., Balding D.J., Guillemaud T., Estoup A. 2008 Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24(23), 2713-2719. (doi:10.1093/bioinformatics/btn514). 6. Storz J.F., Beaumont M.A. 2002 Testing for genetic evidence of population expansion and contraction: An empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution 56(1), 154-166.