Supplementary Information (doc 3324K)
advertisement

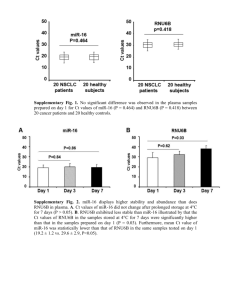
Direct Regulation of microRNA Biogenesis and Expression by Estrogen Receptor Beta in Hormone-responsive Breast Cancer O. Paris et al. Supplementary Information Supplementary Materials and methods Microarray analyses; MicroRNA expression profiling data analysis; Immunohistochemistry. Supplementary References 7 references. Supplementary Tables S1-S4 Supplementary Figures S1-S6 with legends Supplementary Materials and methods Microarray analyses. For miRNA expression profiling, both biological (multiple experiments) and technical (multiple arrays) replicates were performed. In the case of cells lines analyzed in standard growth conditions (complete medium), 800 ng of purified total RNA were fluorescently labelled, amplified and hybridized in duplicate on Illumina v.2 MicroRNA Expression Bead Chips (Illumina Inc.). Also, 100 ng of the same from control cells (MCF-7 wt) and from TAP-ERβ expressing cells were fluorescently labelled, amplified and hybridized in duplicate on Agilent Human microRNA Microarray 18x15K v.3 (Agilent Technologies, Inc.). In each case analyses were performed according to protocols provided by the array manufacturer. For cells arrested at G0-G1 and then stimulated with 17-estradiol (+E2) and for FFPE tumor tissue samples analysis, RNA was fluorescently labelled and amplified in triplicate, pooled and hybridized to Illumina v.2 MicroRNA Expression BeadChips (Illumina Inc.), according to the protocols provided by the array manufacturer. For mRNA expression profiling, 500 ng total RNA from C-TAP-ER, C-TAP-ER and N-TAPER were reverse transcribed, as described previously (Cicatiello et al., 2010; Ravo et al., 2008) and used for synthesis of cDNA and biotinylated cRNA according to the Illumina TotalPrep RNA amplification kit (Ambion; code IL1791) sample preparation kit protocol. For each sample, 700 ng of cRNA were hybridized for 18 hours at 55°C on Illumina HumanWG-6 version 2.0 BeadChips containing 48701 probes (Illumina Inc.), according to the manufacturer’s protocol. The BeadChips was scanned using Illumina BeadArray Reader 500 according to the manufacturer’s standard methods. The microarray data were deposited in the Array Express repository database with the following accession numbers: E-TAMB-1053 for miRNA expression profiling performed in breast cancer cell lines in standard growth conditions, E-TAMB-1055 for miRNA expression profiling performed in starved and hormone-stimulated breast cancer cell liness; E-TAMB-1054 for miRNA expression profiling in FFPE tumor samples; E-TAMB-1052 for mRNA expression profiling in breast cancer cell lines. MicroRNA expression profiling data analysis. For analysis of miRNA expression profiling data generated with Illumina microarrays, the fluorescence intensity files were loaded into the Illumina GenomeStudio v2009.1 software for quality control and expression analysis. First, the quantile normalization algorithm was applied on the raw datasets to correct systematic errors. This normalization equalizes distribution, median and mean of probe intensities among all sample, as the normalization distribution is chosen by averaging each quantile across samples. For differential expression analysis, technical replicates were grouped together and miRNAs with a detection pvalue below 0.01, corresponding to a false positive rate of 1%, were considered as expressed. Statistical significance was calculated with the Illumina DiffScore, a proprietary algorithm that uses the bead standard deviation to build an error model. Only miRNAs with a DiffScore ≤-20 and ≥20, corresponding to a p-value of 0.01, were considered as statistical significant. For miRNA expression profiling generated with Agilent microarrays, raw data were obtained with the Agilent Feature Extraction (AFE) software v.10.7 (Agilent Technologies, Inc.). The AFE algorithms estimate a single intensity measure for each miRNA, referred to as total gene signal (TGS). The AFE-TGS is estimated by multiplying the total probe signal by the number of probes per gene. After getting the raw data, we identified miRNAs differentially expressed in hormoneresponsive human breast cancer cells ERβ+ vs ERβ-, using GeneSpring gx v.11 software (Agilent Technologies, Inc.). We performed data normalization to the 75th percentile; detected miRNAs were then filtered based on their flag values P (Present) present in the data file. The GeneSpring software considers the "gIsGeneDetected" as the flag column and marks microRNA marked with '0' as Absent and '1' as Present. Only miRNAs 'present' in at least one sample were further considered and are displayed. Finally miRNA differential expression between two given conditions was determined calculating ERβ+/ ERβ- ration (fold-change) with a cut-off of ≥1.2. RNA expression profiling data analysis was performed as described earlier (Cicatiello et al., 2010; Ravo et al., 2008). Transcripts were considered detected when the corresponding detection p-value was ≤0.01. Differentially expressed RNAs were selected when DiffScore was ≤-40 or ≥40, values corresponding to a p-value of 0.0001. Raw data were normalized as described above using GenomeStudio v. 2009.1 and analyzed with GeneSpring. Background was subtracted and spots corresponding to absent or low-quality signals were removed from the analysis before global median normalization. Expression data were median-centered by using a per-chip 50th percentile method that normalizes each chip on its median, allowing comparison among chips. miRNAs with a detection p-value below 0.05 were considered as expressed. Hierarchical clustering was performed with the Manhattan algorithm. To identify differentially expressed miRNAs between ER+ and ER- samples, a fold-change cut-off of ±1.2 was applied. Immunohistochemistry. Formalin-fixed paraffin-embedded breast specimens were cut on SuperFrost Plus slides (Menzel-Glaser). Antigen retrieval was performed by microwave at 430 W (1mM citrate buffer, pH6.0) for two cycles of 10 minutes each and one of 5 minutes for anti ER monoclonal antibodies (MoAbs) and by thermostatic bath at 96°C (10 mM/L citrate buffer, pH 6.0) for 40 minutes for ER PgR, HER2 and Ki67. Sections were incubated with the anti-ER-1 MoAb PPG5/10 (GeneTex, dilution 1:15; Skliris et al., 2002) overnight at 4°C, with the anti-ER MoAb 6F11 (Novocastra), the anti-PgR MoAb 1A6 (Novocastra), the anti-Ki67 MoAb MIB-1 (Dako) and the anti-HER2 polyclonal antibody (A0485, Dako) for 30 min at room temperature. Positive and negative controls were included for each antibody and in each batch of staining. The immunoreactions were revealed by a streptavidin-biotin enhanced peroxidase system (Super Sensitive Link-Label IHC Detection System, Biogenex) using 3-amino-9-ethylcarbazole (Dako) as chromogenic substrate. Evaluation of the immunohistochemical results, was performed independently and in blinded manner by two investigators (M.M. and F.N.) using a light microscope (Nikon, Eclipse 55i) equipped with a software to capture images (Eureka Interface System). ER carcinoma cell nuclei, and positive when a moderate/strong staining reaction was observed in 20% to 100% of neoplastic cell nuclei. This cut-off, which was in agreement to Shaaban et al. (2003) and Gruvberger-Saal et al. (2007), was generated using the Classification and Regression Tree (C&RT) analysis. We introduced into the model three variables (nodal status, ER expression, relapses) as described earlier (Novelli et al., 2008). ER and PgR were considered positive when >10% of the neoplastic cells showed a distinct nuclear immunoreactivity whereas Ki67, based on the median value of a referral series of BC patients, was regarded as high if >15% of the cell nuclei were immunostained. HER-2 immunostaining was performed following the manifacturer’s protocol and protein overexpression was determined as defined in the HercepTest kit guide: score 0, 1+, 2+, 3+. All IHC 2+ tumors were analyzed with Chromogenic in situ hybridization (CISH) as previously described (Vocaturo et al., 2006), in order to determine the HER2 gene copy level. Cases with score 0, 1+ and 2+ lacking amplification were considered negative whereas amplified cases with score 2+ and cases with score 3+ were considered positive. Supplementary References Cicatiello L, Mutarelli M, Grober OM, Paris O, Ferraro L, Ravo M, et al. (2010). Estrogen receptor alpha controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and microRNAs. Am J Pathol 176: 2113-2130. Gruvberger-Saal SK, Bendahl PO, Saal LH, Laakso M, Hegardt C, Edén P, et al. (2007). Estrogen receptor beta expression is associated with tamoxifen response in ERalpha-negative breast carcinoma. Clin Cancer Res 13: 1987-1994. Novelli F, Milella M, Melucci E, Di Benedetto A, Sperduti I, Perrone-Donnorso R, , et al. (2008). A divergent role for estrogen receptor-beta in node-positive and node-negative breast cancer classified according to molecular subtypes: an observational prospective study. Breast Cancer Res 10: R74. Ravo M, Mutarelli M, Ferraro L, Grober OMV, Paris O, Tarallo R, et al. (2008). Quantitative expression profiling of highly degraded RNA from formalin-fixed, paraffin-embedded breast tumor biopsies by oligonucleotide microarrays. Lab Invest 88: 430-440. Shaaban AM, O'Neill PA, Davies MP, Sibson R, West CR, Smith PH, et al. (2003). Declining estrogen receptor-beta expression defines malignant progression of human breast neoplasia. Am J Surg Pathol 27: 1502-1512. Skliris GP, Parkes AT, Limer JL, Burdall SE, Carder PJ, Speirs V (2002). Evaluation of seven oestrogen receptor beta antibodies for immunohistochemistry, western blotting, and flow cytometry in human breast tissue. J Pathol; 197:155-62. Vocaturo A, Novelli F, Benevolo M, Piperno G, Marandino F, Cianciulli AM, , et al. (2006). Chromogenic in situ hybridization to detect HER-2/neu gene amplification in histological and ThinPrep-processed breast cancer fine-needle aspirates: a sensitive and practical method in the trastuzumab era. Oncologist 11: 878-886. Supplementary Table S1. Relationship between ERβ expression and bio pathological variables Variable Histotype Invasive ductal carcinoma Invasive lobular carcinoma pT T1 T2 T 3-4 pN Negative Positive G 1-2 3 ERα Negative (≤10%) Positive (>10%) PgR Negative (≤10%) Positive (>10%) HER2 Negative (0/1+) Positive (2+/3+) Ki-67 Low (≤15%) High (>15%) N° of cases (%) ERβ+ (%) ERβ- (%) p-value 1 37 (92.5) 3 (7.5) 20 (54) 2 (67) 17 ( 46) 1 (33) 0.673 28 (70) 9 (22.5) 3 (7.5) 18 (64) 3 (33) 1 (33) 10 (36) 6 (67) 2 (67) 0.197 27 (67.5) 13 (32.5) 21 (78) 1 (8) 6 (22) 12 (92) <0.0001 29 (72.5) 11 (27.5) 19 (66) 3 (27) 10 (34) 8 (73) 0.03 3 (7.5) 37 (92.5) 2 (67) 20 (54) 1 (33) 17 (46) 0.67 9 (22.5) 31 (77.5) 6 (67) 16 (52) 3 (33) 15 (48) 0.43 28 (70) 12 (30) 15 (54) 7 (58) 13 (46) 5 (42) 0.78 24 (60) 15 (62.5) 16 (40) 7 (44) 9 (37.5) 9 (56) 0.24 pT: Tumor size; pN: Lymphnodes; G: Grading; ERα: Estrogen Receptor alfa; ERβ: Estrogen Receptor beta; PgR: Progesterone Receptor. 1 2 test Supplementary Table S2. Sixty-seven miRNAs differentially expressed in ER positive vs in ER negative breast tumors miRNA Foldchange p-value miRNA 1,55 -1,23 -1,36 -1,76 1,61 -1,50 1,30 1,31 1,51 -2,47 0,03300 0,01330 0,04961 0,00478 0,00491 0,00436 0,01662 0,01692 0,00260 0,01408 HS_76 hsa-miR-106a:9.1 hsa-miR-1248 hsa-miR-1273 1,37 -1,45 1,60 -1,21 3,65 0,01582 0,02278 0,03216 0,04955 0,01661 hsa-miR-1285 hsa-miR-129* hsa-miR-1300 hsa-miR-1301 hsa-miR-1304 hsa-miR-1321 hsa-miR-138-1* hsa-miR-146b-5p hsa-miR-155 hsa-miR-181b hsa-miR-19b hsa-miR-200b* hsa-miR-202 hsa-miR-21* hsa-miR-216a hsa-miR-300 hsa-miR-30a hsa-miR-30a* hsa-miR-323-3p 2,20 -1,45 1,63 -1,34 1,90 -1,30 1,89 -1,24 -1,29 -1,22 1,24 1,23 1,23 -1,26 1,51 1,30 1,25 1,47 -1,75 0,04450 0,01608 0,01434 0,01527 0,01300 0,02943 0,02667 0,01200 0,04829 0,02224 0,03712 0,01470 0,03878 0,04976 0,04616 0,00264 0,04286 0,03410 0,01749 HS_108.1 HS_1341 HS_170 1 HS_240 1 HS_251.1 1 HS_266.1 HS_276.1 HS_286_a (hsa-miR-758) HS_40 HS_44.1 HS_69 a a 1 Foldchange p-value hsa-miR-329 hsa-miR-337-5p hsa-miR-342-5p hsa-miR-34b hsa-miR-367 hsa-miR-409-3p hsa-miR-424* hsa-miR-431 hsa-miR-450b-3p hsa-miR-454* -1,57 -1,48 1,33 -1,64 1,20 -1,45 -1,56 -2,16 -1,36 -2,09 0,04835 0,03259 0,00626 0,02189 0,01750 0,03871 0,03287 0,00322 0,04701 0,01347 hsa-miR-487b hsa-miR-491-5p hsa-miR-499-3p hsa-miR-505 hsa-miR-518e hsa-miR-518f/d-5p/520c5p/526a hsa-miR-520d-3p hsa-miR-548d-3p hsa-miR-616 hsa-miR-616* hsa-miR-625* hsa-miR-662 hsa-miR-663b hsa-miR-7 hsa-miR-708* hsa-miR-801:9.1 hsa-miR-873 hsa-miR-886-3p hsa-miR-922 hsa-miR-92b* hsa-miR-941 solexa-2526-361 solexa-7764-108 -1,46 1,32 1,32 -1,43 -1,28 0,03464 0,04791 0,03770 0,01903 0,04002 1,36 1,61 -1,28 -1,42 -1,44 1,43 1,41 -1,57 -1,51 -1,50 1,45 1,49 -1,65 -1,22 -1,60 -1,46 1,52 2,33 0,01679 0,01742 0,03891 0,01638 0,02911 0,02250 0,04948 0,04488 0,01951 0,01346 0,04721 0,02599 0,03621 0,01549 0,02824 0,02612 0,02596 0,00998 The miRNA probe in the array matches multiple loci in the genome Bold: miRNAs differentially expressed also in ER–positive vs ER–negative hormone-responsive human breast cancer cells in culture Supplementary Tables S3. Estrogen receptor binding sites within 10kb from loci encoding ER-responsive miRNAs S3-A miRNA differentially expressed in ER+ vs ERcells Closest ER binding site upstream1 Closest ER binding site downstream1 Closest ER binding site upstream1 Closest ERbinding site downstream1 Closest ER binding site upstream1 ER+ cells Closest ER binding site downstream1 ER- cells 0 (overlapping) HS_166.1 – – – – – HS_266.1 0 (overlapping) 0 (overlapping) 0 (overalapping) 0 (overlapping) (chr17: 5232117-52323792) (chr17: 5232117-52323792) (chr17:523287-52324146) (chr17:523287-52324146) – – – – – – -5986 – – – – – – -5498 – -5459 – – – – – – hsa-let-7a (hsa-let7a-3 gene) hsa-miR-1257 hsa-miR-1285 (miR-1285-2 gene) hsa-miR-181c* hsa-miR-23a* hsa-miR-27a* hsa-miR-24-2* hsa-miR-23b* hsa-miR-27b* hsa-miR-24-1* / hsa-miR-189:9.1 hsa-miR-30a* (chr2:70339139-70339830) -3032 (chr19:13843046-13843481) – (chr2:70339100-70339829) -2998 (chr19:13842946-13843515) (chr22:44880878-44881307) (chr11:64368360-64368992) 2970 (chr20:59958529-59959027) -7039 401 -7082 302 -6791 191 (chr19:13815512-13815956) (chr19:13807463-13808000) (chr19:13815555-13816119) (chr19:13807490-13808099) (chr19:13815264-13816060) (chr19:13807532-13808210) -7181 254 -7224 155 -6933 44 (chr19:13815512-13815956) (chr19:13807463-13808000) (chr19:13815555-13816119) (chr19:13807490-13808099) (chr9: 13815264-13816060) (chr19:13807532-13808210) -7939 101 -7382 2 0 (overlapping) 0 (overlapping) (chr19:13815512-13815956) (chr19:13807463-13808000) (chr19:13815555-13816119) (chr19:13807490-13808099) (chr19:13807532-13808210) (chr19:13807532-13808210) -5704 4878 -5667 4698 -5716 4437 (chr9:96881097-96881607) (chr9:96892285-96893483) (chr9:96881256-96881644) (chr9:96892105-96892943) (chr9:96880936-96881595) (chr9:96891841-96892253) -5941 4641 -5904 4461 -5953 4197 (chr9:96881097-96881607) (chr9:96892285-96893483) (chr9:96881256-96881644) (chr9:96892105-96892943) (chr9:96880936-96881595) (chr9:96891841-96892253) -6517 4094 -6480 3914 -6529 3650 (chr9:96881097-96881607) (chr9:96892285-96893483) (chr9:96881256-96881644) (chr9:96892105-96892943) (chr9:96880936-96881595) (chr9:96891841-96892253) 0 (overlapping) 0 (overlapping) (chr6:72169848-72170426) (chr6:72169848-72170426) – – – – hsa-miR-32* hsa-miR-330-5p hsa-miR-338-3p hsa-miR-365 – -2139 (chr19:50836324-50836783) -9728 (chr17:76724072-76724572) -946 (miR-365-2 gene) (chr17: 26925148-26925597) hsa-miR-548d3p (miR-548d-2 – gene) hsa-miR-616* hsa-miR-663b hsa-miR-935 – 9615 – (chr9:110838378-110838715) 9586 (chr9:110838112-110838744) – – – – – – – – – – – – -743 – -936 – – (chr17:26924937-26925800) 3158 – (chr17:62894141-62894909) – -2852 (chr12:56202161-56202718) 3280 (chr17:62894161-62894787) – (chr17:26925077-26925607) – -2791 (chr12:56202100-56202671) 3304 (chr17:62894122-62894763) – -531 823 -2027 603 -931 811 (chr2:132731654-132732045) (chr2:132729595-132730186) (chr2:132733150-132733597) (chr2:132729599-132730406) (chr2:132732054-132732613) (chr2: 132729555-132730198) – – – 252 – Bold: miRNAs differentially expressed in ER+ vs ER- breast tumors 1 Distance in bps (chr19:59177715-59178175) 361 (chr19:59177824-59178301) S3-B miRNA differentially expressed in ER+ vs ERbreast tumors HS_266.1 hsa-miR-1285 (hsa-miR-1285-2 gene) hsa-miR-129* (hsa-miR-129-1 gene) hsa-miR-181b hsa-miR-21* hsa-miR-30a Closest ER binding site upstream1 0 (overlapping) Closest ER binding site downstream1 ER+ cells 0 (overlapping) 0 (overlapping) (chr17:5232117-52323792) (chr17:5232117-52323792) -5498 – (chr2:70339139-70339830) Closest ER binding site upstream1 – – – – Closest ERbinding site downstream1 0 (overlapping) – – – – – – – – -320 – – (chr17:523287-52324146) (chr17:523287-52324146) -5459 (chr2:70339100-70339829) (chr1:197095054-197095581) 3028 (chr7:127638260127638719) – 0 (overlapping) 0 (overlapping) 0 (overlapping) 0 (overlapping) 0 (overlapping) 0 (overlapping) (chr17:55273240-55273891) (chr17:55273240-55273891) (chr17:55272893-55273703) (chr17:55272893-55273703) (chr17:55273348-55273732) (chr17:55273348-55273732) 0 (overlapping) 0 (overlapping) (chr6:72169848-72170426) (chr6:72169848-72170426) – – – – – – – – -839 – 0 (overlapping) 0 (overlapping) (chr6:72169848-72170426) hsa-miR-342-5p – – – – hsa-miR-409-3p – – – – hsa-miR-487b – – – – hsa-miR-520d-3p – – – – 3158 – hsa-miR-548d-3p Closest ER binding site downstream1 ER- cells (chr6:72169848-72170426) hsa-miR-30a* Closest ER binding site upstream1 (hsa-miR-548d-2 gene) – hsa-miR-616 – chr17:62894141-62894909) – -2855 (chr12:56202161-56202718) 3280 (chr17:62894161-62894787) – (chr14:99644461-99644906) -9836 (chr14:100591066-100591554) – -2535 (chr19:58912177-58912627) – -2791 (chr12:56202100-56202671) – 8438 (chr14:100591066-100591554) – 3304 (chr17:62894122-62894763) – -2855 – – hsa-miR-625* – – – hsa-miR-663b -531 823 -2027 (chr2:132731654-132732045) (chr2:132729595-132730186) (chr2:132733150-132733597) (chr2:132729599132730406) hsa-miR-922 – – – – hsa-miR-92b* – – – – – -6039 – -5975 – – – – – – – – – hsa-miR-941-1 hsa-miR-941-2 hsa-miR-941-3 (chr20:62014658-62015199) -6343 (chr20:62014658-62015199) -6458 (chr20:62014658-62015199) – – – -2791 hsa-miR-616* (chr12:56202161-56202718) – 603 (chr20:62014664-62015263) -6282 (chr20:62014664-62015263) -6394 (chr20:62014664-62015263) Bold: miRNAs regulated by ER in hormone-responsive human breast cancer cells in culture 1 Distance in bps (chr12:56202100-56202671) -118 (chr14:65007006-65007455) – – -931 811 (chr2:132732054-132732613) (chr2:132729555-132730198) -8011 – (chr3:198893855-198894243) 7901 (chr1:153439588-153440193) Supplementary Table S4. Primers used for qPCR analyses ERβ binding Site Chr Forward Primer (5’-3’) Coordinates Reverse primer (5’-3’) Coordinates ERG7838/39 6 TTGAAGTCCGAGGCAGTAGG 72169957-77 ATCCTCGACTGGAAGCTGTG 72170033-14 ERG9242 9 TGCCACTTCAGCACTTCAAA 96881191-210 CCCTCTGAGAACGGGTGTTA 96881278-59 ERG9243 9 CAGCCCCAGGACTTAAAGG 96894350-69 TGGGAGGCTCTGGTCTTTAC 96894411-392 pS2/TFF1 21 CTAGACGGAATGGGCTTCAT 42660162-43 GCTTGGCCGTGACAACAGTG 42660075-94 pri-miR Chr Forward Primer (5’-3’) Coordinates Reverse primer (5’-3’) Coordinates TTCAGCTTTGTAAAAATGTATCAAAGAG AT TGCACCTGTTCTCCAATCTGC 72169875-904 Coordinates Reverse primer (5’-3’) Coordinates pri-miR-30a 6 pri-miR-23b∼24-1 9 pre-miR ATTGCTGTTTGAATGAGGCTTCAGTACTT 72170175-46 T TCACATTGCCAGGGATTACCA 96887369-89 Chr Forward Primer (5’-3’) 96887459-39 pre-miR-30a 6 ATCCTCGACTGGAAGCTGTG 72170033-14 CTGCAAACATCCGACTGAAA 72169979-98 pre-miR-23b 9 AGGTGCTCTGGCTGCTTG 96887314-31 GTCGTGGTTGCGTGGTAA 96887401-384 pre-miR-24-1 9 CTCCGGTGCCTACTGAGC 96888124-41 TCCTGTTCCTGCTGAACTGA 96888190-71 pre-miR-27b 9 TGCAGAGCTTAGCTGATTGG 96887563-82 GTGAACAAAGCGGAAACCA 96887612-594 RPLP0 15 GGCGACCTGGAAGTCCAACT 119122952-33 CCATCAGCACCACAGCCTTC 119121560-79 Supplementary Figure Legends Supplementary Figure S1. Heatmap summarizing the effects of the estrogen responsive transcriptome of MCF-7 cell clones, showing changes in mRNA expression (fold-change) of cell clones grown in standard culture condition. Supplementary Figure S2. (A) Real-Time PCR validation of 10 miRNAs identified by microarraya analysis in wt, N-TAP-ERβ and C-TAP-ERβ cells. (B) Correlation graph between fluorescence intensity log2 value from microarray data and real-time rtPCR log2 data in ERβ+ and ERβ- cells. Supplementary Figure S3. (A) Heatmap and (B) Graphics showing miRNAs miR-30a* and miR23b*/-27b*/-24-1* fluorescence levels compared with those of their complementary strand in CTAP-ERα and TAP-ERβ cells upon stimulation with E2 for the indicated times. In panel (A) fluorescence levels are reported as log2. Supplementary Figure S4. Functional analysis according to Gene Ontology of predicted mRNA targets of miRNAs differentially expressed between ERβ+ and ERβ- cells according to miRBase (release 12). Supplementary Figure S5. Immunohistochemistry of representative BC biopsies. (A) ERβ negative, (B) and (C) ERβ positivity: 30-50%, (D) ERβ positivity: 80-90%. Supplementary Figure S6, (A) Genome Browser view of ERβ and ERα binding sites within 10 kb upstream or downstream from miR-23b/27b/24-1 cluster and ORF (c9orf3) in which the cluster is present in Chromosome 9. (B) and (C) Validation of ERβ binding sites by ChIP and real time PCR in wt, N-TAP-ERβ and C-TAP-ERβ cells stimulated for 45 min with 10-8M E2. Figure S1 Figure S2 Figure S3 Figure S4 Figure S5 Figure S6