Lecture Three - Personal Webspace for QMUL
advertisement

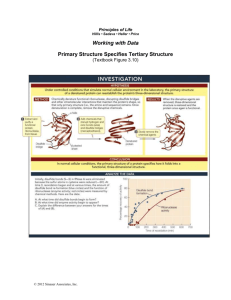
Lecture Three: Protein Structure contd…. [Chapter 2 – Berg, Tymoczko & Stryer] (Figures in Red are for the 7th Edition) Primary Structure The _____________ (and disulphides) Secondary Structure Local sequence conformation Particularly repeating units -helix -pleated sheet DEFINITION The Tertiary Structure of a Protein The spatial arrangement of amino acids usually far apart from each other in the linear sequence Myoglobin (Figure 2-48 pages 47) (2-43, page 46) and Concanavalin A These are example proteins consisting of ______ ______________ chains Each has a specific structure DEFINITION The Native Structure of a Protein The three dimensional structure of a protein when in a _______________________ (Figure 6-3, page 166) (6-3, page 183) Ribonuclease A (RNase A) The structure of RNase A comprises -helices and -Sheets 124 amino acids ________ disulphide bonds In the native structure the correct arrangement is 26-84 40-95 58-110 67-72 RNase A hydrolyses RNA Experiments with RNase A determined the key factors involved in producing a native structure Christian Anfinsen’s Experiments Background to Experiment In a solution of either Urea (8M) or Guanidine Hydrochloride (6M) proteins lose their _______________ Proteins Denature Unfolded protein - Random Coil -Mercaptoethanol is a _______________ which breaks disulphide bonds Experiment One Figure 2-58, page 51 (2-53, page 49) Dissolve RNase A in a solution of -Mercaptoethanol and 8M Urea Result: The RNase A completely loses enzyme activity Denatured RNase A Then remove the -Mercaptoethanol and 8M Urea Involves a procedure called _________ Oxidise the cysteine residues to _______ disulphide bonds Result: The RNase A spontaneously regains all its enzyme activity Renatured RNase A Significant Conclusion The amino acid sequence of RNase A provides the information needed to specify its native structure THIS IS TRUE FOR ALL PROTEINS The Primary Structure of a protein dictates its Tertiary Structure Experiment Two Figure 2-58 & Figure 2-59, pages 51 & 52 (2-53, p.49, 2-54, p.50) Denatured RNase A Initially remove ONLY the -Mercaptoethanol but RETAIN the 8M Urea Result: RNase A slowly regains ~1% enzymatic activity Reason: There is only one correct arrangement of disulphides but 105 different combinations 1/105 1% Now add trace amounts of -Mercaptoethanol The disulphide bonds can _________ Result: After ~10 hours RNase A completely regains its enzymatic activity ________________ has driven the protein to its native structure Further Significant Conclusion The thermodynamically most stable structure of RNase A is its ______________ Also true for all proteins Footnote: Anfinsen obtained the 1972 Nobel Prize for Chemistry for his work with RNase A DEFINITION The Quaternary Structure of a Protein The spatial arrangement in a protein made up from more than one polypeptide chain Each chain is called a _________ of the protein They are referred to by either Greek or Alphabetic letters Haemoglobin (Look also at Figure 2-54, page 50) (2-49, p.48) Haemoglobin is an example protein with quaternary structure It has FOUR separate polypeptide chains Two identical alpha subunits Two identical beta subunits The structure is an 2 2 configuration Reasons for quaternary structure Enables smaller quantities of genetic material to create larger proteins and structures Example: Viruses use 60 repeats of a four subunit structure to produce a highly symmetric coat Quaternary structure is excellent for _____________ ____________ This is through the allosteric effect A change in one subunit induces a change in another DEFINITION Protein Conformation The three-dimensional arrangement of a proteins atoms in its structure Protein conformation is independent of the number of chains in the protein A protein is ______________ in its native conformation Protein folding The principal factor governing protein folding is the burying of ____________ side chains This forms the Protein Core Levinthal’s Paradox Levinthal calculated that a protein containing 100 residues would take 1.6 x 1027 YEARS to fold to a native structure by using random chance at getting the correct structure RNase A folds in seconds to its native structure Conclusion: Proteins do not fold via a random pathway Answer: Proteins fold to their native structures via the formation of stable partially correct secondary structure features as intermediate stages in the folding process This results in proteins taking only seconds to form their native structures Summary of Lecture Three: The Tertiary Structure of a protein is the conformation adopted by a single polypeptide chain The Quaternary Structure of a protein is the conformation adopted by a protein containing more than one polypeptide chain Protein folding buries hydrophobic residues The Protein Core Proteins fold via the formation of stable partially correct secondary structure intermediate stages