Supplementary Material (doc 32K)
advertisement

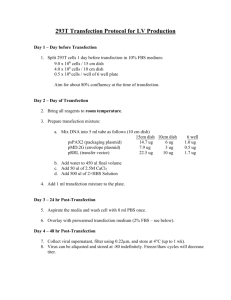
SUPPLEMENT MATERIAL MS # ONC 2006-00309RR Title: RalA regulates vascular endothelial growth factor-C (VEGF-C) synthesis in prostate cancer cells during androgen ablation Author: Francesca Rinaldo, Jinping Li, Enfeng Wang, Michael Muders, and Kaustubh Datta Detail methodology for Real-time PCR: Real-time PCR was performed according to the Taqman method in a 25 L volume using 1 g total RNA, 12.5 l RT-PCR Master Mix (Applied Biosystems), 0.6 l RNase inhibitor (Applied Biosystems), 50nM forward and reverse primers and 100 nM middle primer. An ABI System Sequence Detector 7700 (Applied Biosystems) was used with the following regimen of thermal cycling: Stage 1: 1 cycle, 30 minutes at 48˚ C; Stage 2: 1 cycle, 10 minutes at 95˚ C; Stage 3: 40 cycles, 15 seconds at 95˚ C, 1 minute 60˚ C. Relative mRNA amounts were calculated as follows. The cycle number (CT) at which PCR amplification took place for a particular threshold value in the exponential phase for each reaction was determined by the sequence detector. Real-time PCR for the house keeping gene, 36B4 was performed for each test sample along with VEGF-C. To normalize the value of VEGF-C for each reaction condition, the value of 36B4 at that condition was deducted and the resulting value was designated as ∆. Therefore, ∆=CT (VEGF-C sample)-CT (36B4 sample). To calculate the relative expression of the treated sample as compared to the control sample, first was determined by deducting the value of the control sample from the treated sample. Therefore, ∆∆=∆ (transfected or treated sample) - ∆ (empty vector or untreated sample). Finally, relative RNA amount in comparison to the control was calculated by using the formula, 2-. Average and standard deviation for three experiments were calculated. P values for statistical significance were also calculated. Cell Transfections and RalA siRNA: LNCaP cells were plated in 60 mm cell culture dishes 24 hours prior to transfection (approximately 5 x 105 cells per dish). The Effectene transfection reagent kit (Quiagen) was used to transfect the RalN28 (PMT2-HA-Ral-N28) dominant negative expression vector, the JNK1 (PCDNA3-Flag-JNK1) dominant negative expression vector and to generate retrovirus carrying the RalA Q75L (pBabe-puro-RalA-Q75L) dominant active expression vector. Human dominant negative RalN28 expression vector was kindly provided to us by Dr. Johannes L. Bos. JNK1 dominant negative expression plasmid was a kind gift from Dr. Roger J Davis. The RalA Q75L expression vector was provided to us by Dr. Channing J Der. PMT2 and PCDNA3 empty vectors were used as controls for transfection experiments; LacZ retrovirus was used as a control for cell infection experiments. All transfections were carried out according to the Quiagen Effectene transfection reagent kit protocol. In preparation for RalA siRNA transfection, LNCaP cells were cultured in 10% FBS or Charcoal Stripped FBS for 24 hours at 37.4˚ C. RalA Duplex I (Dharmacon RNA Technologies) was diluted to a concentration of 20 M in 1X universal buffer (20 mM KCl, 6mM HEPES pH 7.5, 0.2 mM MgCl2). Transfection of LNCaP cells with 200 nM RalA siRNA duplex was achieved using the TransIT-SiQuest Transfection Reagent (Mirus). siRNA transfection was allowed to proceed 72 hours before collection of whole cell extract or total RNA as outlined below. Target sequence: 5’-AAG ACA GGT TTC TGT AGA AGA-3’. RNA Duplex: 5’- AAG ACA GGU UUC UGU AGA AGA dTdT dTdT UUC UGU CCA AAG ACA UCU UCU- 5’.