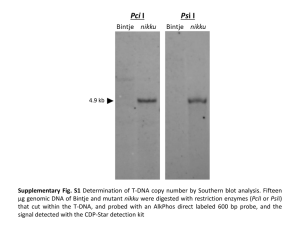
Supplementary Information For Homology Modeling, Molecular
advertisement

Supplementary Information For Homology Modeling, Molecular Dynamics, e-Pharmacophore mapping and Docking Study of Chikungunya Virus nsP2 Protease Kh.Dhanachandra Singh1¥, Palani Kirubakaran 1¥, Shanthi Nagarajan2¥, Sugunadevi Sakkiah3, Karthikeyan Muthusamy1, Devadasan Velmurgan5*, Jeyaraman Jeyakanthan1*. 1 Department of Bioinformatics Alagappa University, Karaikudi – 63003, India 2 Bioinformatics Centre, Pondicherry University, Pondicherry - 605 014, India 3 Bioinformatics Centre and Computational Biology Laboratory, Department of Bioinformatics, Bharathiar University, Coimbatore - 641 046, India 5 CAS, Biophysics and Crystallography, University of Madras, Chennai - 600025, India ¥ These authors contributed equally to this work. Supplementary Fig.1. Ramachandran plot of nsP2 protease model built using NCBI Structural Analysis and Verification server. Supplementary Fig.2. ProSA energy plot calculated for nsP2 protease model. Supplementary Fig.3. Graph showing the total energy (a), potential energy (b), pressure (c), temperature (d) and volume (e) against the simulation time of 5 ns. Supplementary Fig.4. Multiple sequence alignment of nsP2 protease sequences reported in UniProt from Chickungunya virus. Mutation was not found in the active site residues reported.