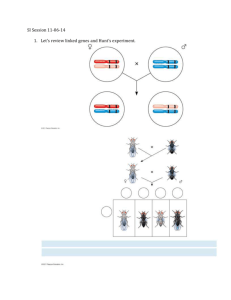
Linkage Mapping in the Peppered Moth Positions
advertisement

1 1 SUPPLEMENTARY INFORMATION 2 Figure S1 Quality improvement of a co-dominant AFLP. mCCeACC171- 3 175 was a co-dominant AFLP marker with two distinct alleles of 171 and 175 4 bp, but with a pattern that is somewhat ambiguous. The original AFLP traces to 5 the left of the vertical black bar were generated with the Mse+CC primer, 6 whereas the traces to the right of the bar are for the same individuals amplified 7 with Mse+CCAA, which was used to generate a reliable genotype 8 (mCCAAeACC171-175 marker on LG25). 9 10 Figure S2 11 extension. The bottom trace shows a region of the original Hha+GA trace with 12 18 peaks, including a 222 bp peak that was selected for sequencing. The four 13 traces above have alternative 3-base (GAN) extensions. The original peaks 14 separate into the four groups, except for the peak indicated with the blue line, 15 which is an overlapping peak in the +2 trace (size homoplasy), and splits into 16 two peaks in the +3 profiles. The +3 peaks are higher (vertical scales are not 17 identical) and more isolated. Separation of a complex AFLP trace by selective primer 18 19 Figure S3 Basic cytogenetic characteristics of Biston betularia. (a) DAPI- 20 stained mitotic spermatogonial metaphase showing 2n = 62 chromosomes; (b) 21 YOYO-1 stained spermatocyte complement showing n = 31 meiotic metaphase 22 I bivalents (individual bivalents are arbitrarily numbered to facilitate their 2 23 distinction); (c) DAPI-stained male pachytene complement showing n = 31 24 meiotic bivalents (chromomere pattern is not seen due to the pretreatment in 25 hypotonic solution); (d) DAPI-stained female pachytene complement (n = 31) 26 showing chromomere patterns; note the WZ bivalent is easily identified from 27 the DAPI-highlighted heterochromatic thread of the W chromosome; (e) 28 Orcein-stained polyploid nucleus of the female Malpighian tubule cell showing 29 a large spherical sex-chromatin body (arrow), representing multiple copies of 30 the heterochromatic W chromosome; (f) orcein-stained polyploid nucleus of the 31 male Malpighian tubule cell without sex chromatin. Scale bars indicate 5 μm in 32 (a, b), 10 μm in (c, d), and 20 μm in (e, f). 33 34 Figure S4 Gene order rearrangements between B. betularia and B. mori. 35 Gene order rearrangements were detected between B. betularia and B. mori in 36 five linkage groups. Two of these are shown in this figure, the others are 37 included in Figure 3 (LG11 and LG29) and in Figure 4 (Z). The B. mori 38 chromosomes are scaled according to scaffold sizes in SilkDB using estimated 39 gap sizes, which differ slightly from the fixed gap sizes in Kaikobase. 40 41 Figure S5 Reconstruction of Z chromosome rearrangements between 42 Biston betularia and Bombyx mori. Hypothetical scheme of rearrangements, 43 which could explain the different gene order that differentiated the Z 44 chromosomes of Bombyx mori (left) and Biston betularia (right) from a common 3 45 ancestor. Double-headed arrows indicate inversions, single-headed arrows 46 transpositions; colour of arrows corresponds to colour of repositioned genes. 47 For simplicity, unidirectional alterations from B. mori to B. betularia (from left to 48 right) are illustrated; however, as the ancestral structure of the Z chromosome 49 is unknown, reverse unidirectional alterations (from right to left) or bidirectional 50 alterations from any intermediate structure are equally possible. 51 52 Table S1 Blast results and primers for the mapped genes. Details of the 53 genes included in the linkage map: gene identity, GenBank accession number, 54 NCBI and SilkDB blast results and primers used to isolate and genotype the 55 genes. The SilkDB blast results present the best blastx hit, unless tblastx 56 produced a better hit that was not annotated. The tblastx hits have no Bombyx 57 gene identifier. 58 59 Table S2 Paternal segregation pattern of mapping family (‘16’) offspring. 60 The alleles have been translated from SNP nucleotides, indel large/small, or 61 AFLP present/absent into a binary 1 or 0 code representing the chromosomal 62 origin of the allele. "md" stands for missing data. Conditional formatting is used 63 to demonstrate the recombination breakpoints per individual (vertical 64 orientation) by means of a green/white change. For example, individual 1 does 65 not have any recombination over the full length of LG1, while individual 2 66 recombined between 6-PGD and tpi. 4 67 68 Table S3 69 Bombyx mori). Gene names are listed in the first column with alternative 70 identifiers or frequently used abbreviations in brackets. Methods used to 71 confirm Z-linkage are abbreviated as: FISH (fluorescence in situ hybridization); 72 South (Southern hybridization); LM (linkage mapping); Inh (hemizygous 73 inheritance). References are numbered in square brackets and listed in full 74 below the table; all B. betularia data originate from this study. Only confirmed 75 Z-linked genes are given in O. nubilalis and only the two genes specifically 76 mentioned in the text are included for P. xylostella since only uniref90 best hits 77 are available for this species. All except two genes (31/33) are also Z-linked in 78 B. mori. MYST histone acetyltransferase (BGIBMGA006995) is located on 79 chromosome 17 in B. mori and the identity of Papilio spp. acid phosphatase is 80 unclear; however no acid phosphatase homologues occur on the Z- 81 chromosome scaffolds of B. mori. 82 List of published lepidopteran Z-linked genes (excluding