Supplementary tables

Supplementary Information
Supplementary Methods
Immunohistochemistry
Slides from paraffin-embedded tissue were incubated in xylene for 10 minutes followed by a 100% ethanol wash for 5 minutes, a 75% ethanol wash for 3 minutes and a 50% ethanol wash for 3 minutes. Slides were then rinsed with 1X phosphate buffered saline (PBS) before being placed into citrate buffer for 1 hour at 85°C. This was followed by an incubation in 0.2% Triton X-100/PBS for 30 minutes and a blocking step with 10% NGS in 0.02% Triton X-100/PBS for 1 hour. A rabbit polyclonal antibody was purchased from Abcam Inc and used at a dilution of 1:200 in 2% NGS in 0.02% Triton X100/PBS overnight at 4°C. A secondary antibody, Alexa Fluor 594 (Invitrogen; 1:500), was used in 2% NGS in
0.02% Triton X-100/PBS for 1 hour at room temperature. The slides were finally mounted with VectaShield (Vector Laboratories) mounting medium with DAPI.
Protein extraction and fractionation
Lymphoblast cell lines were derived using standard methods from human whole blood extracted from patients and their unaffected relatives. In 6-well dishes,
MG-132 was added to select wells to achieve a final concentration of 20
M for 6 hours. Cells were transferred to 15 mL falcon tubes and pelleted by centrifugation at 1500 rpm for 5 minutes at room temperature followed by a PBS wash under the same conditions, and a subsequent PBS wash and centrifugation
in 1.5 mL eppendorf tubes. The pellet was re-suspended in 25
l of 0.2% Triton-
X, chilled on ice for 20 minutes and then sonicated at setting 3 for five 0.5 second pulses. Following this the tubes were centrifuged at maximum speed for 15 minutes at
4°C. The supernatant then contained the soluble fraction, while the insoluble pellet was re-suspended in 25
l SUB buffer.
Western blot
A total of 15 µg of each protein sample was resolved on a 12% polyacrylamide gel and transferred to a Nitrocellulose membrane. Membranes were blocked in
0.1% PBS-Tween with 5% BSA for one hour followed by incubation with primary antibody (rabbit polyclonal RNF170; Abcam) at a dilution of 1:100 in 0.1% PBS-
Tween with 5% BSA overnight at 4 o C. Following washing, the membranes were incubated in donkey anti-rabbit HRP (Jackson ImmunoResearch; 1:10,000) in
0.1% PBS-Tween with 5% BSA 1 hour at room temperature. An ECL kit was used for detection followed by a 10-second exposure.
Similar methods were used for detection of actin. However, HRP was inactivated using 0.1% sodium azide solution in 0.1% PBS-Tween with 5% milk. The primary antibody was mouse anti-actin (Miilipore; 1:20,000) added in mixture with sodium azide for 1 hour, while the secondary antibody was donkey anti-mouse
HRP (Jackson ImmunoResearch; 1:25,000) in 0.1% PBS-Tween with 5% milk for one hour at room temperature.
Bioinformatic prediction programs used to assess mutation severity
The effect of amino acid substitution on protein function was predicted with SIFT
(Ng 2001, Ng 2002, Ng 2003), PolyPhen (Sunyaev 2000, Sunyaev 2001,
Ramensky 2002), PANTHER (Thomas 2003, Thomas 2004), and Align-GVGD
(Tavtigian 2006, Mathe 2006). The protein sequence of RNF170 was used as the input sequence for SIFT, PolyPhen, and PANTHER. Query options used for
PolyPhen prediction are Structural database: PQS; Sort hits by: Identity; Map to mismatch: No; Calculate structural parameters: For all hits; Calculate contacts:
For all hits; Minimal alignment length: 100; Minimal identity in alignment: 0.1;
Maximal gap length in alignment: 50; Threshold for contacts: 6Å.
Multispecies alignment
Homologous protein sequences of the human RNF170 gene were retrieved from
NCBI genome database with BLASTP. The multiple sequence alignments MSA 1 and 2 were generated with MUSCLE (Edgar 2004). Supplemental figures 3 and 4 were generated with GeneDoc. The sequence logo was created using WebLogo
(Crooks 2004). Sequences used for MSA2 (including more distant species):
>gi|237858654|ref|NP_112216.3| RING finger protein 170 isoform a [Homo sapiens]
>gi|76655999|ref|XP_874720.1| PREDICTED: RNF170 protein-like [Bos taurus]
>gi|149057838|gb|EDM09081.1| similar to ring finger protein 170, isoform CRA_c [Rattus norvegicus]
>gi|148692281|gb|EDL24228.1| mCG1050561 [Mus musculus]
>gi|295148228|ref|NP_001171208.1| RING finger protein 170 [Gallus gallus]
>gi|62857749|ref|NP_001016872.1| RING finger protein 170 [Xenopus (Silurana) tropicalis]
>gi|50726886|ref|NP_999915.1| RING finger protein 170 [Danio rerio]
>gi|47210826|emb|CAF90883.1| unnamed protein product [Tetraodon nigroviridis]
>gi|115670889|ref|XP_783372.2| PREDICTED: hypothetical protein [Strongylocentrotus purpuratus]
>gi|156364925|ref|XP_001626594.1| predicted protein [Nematostella vectensis]
>gi|170590886|ref|XP_001900202.1| putative LAG1-interacting protein [Brugia malayi]
>gi|17537047|ref|NP_496760.1| hypothetical protein Y38F1A.2 [Caenorhabditis elegans]
>gi|21554064|gb|AAM63145.1| unknown [Arabidopsis thaliana]
>gi|115456477|ref|NP_001051839.1| Os03g0839000 [Oryza sativa Japonica Group]
>gi|56754722|gb|AAW25546.1| SJCHGC08969 protein [Schistosoma japonicum]
>gi|167523711|ref|XP_001746192.1| hypothetical protein [Monosiga brevicollis MX1]
Supplemental results
Bioinformatic prediction of the effect of the R199C missense mutation
The missense mutation R199C was predicted to have deleterious effect on protein function by all four methods SIFT, PolyPhen, PANTHER, and Align-
GVGD.
Using the program SIFT which incorporates sequence homology, the normalized probability for the substitution of R199 to C is calculated to be 0.00, which is less than the threshold 0.05. Thus the mutation is predicted to be deleterious. In the
PolyPhen program, the PSIC ( P ositionS pecific I ndependent C ounts) score difference for the two amino acids R and C is 2.223 (>2.0). The variant is predicted to be probably damaging. The number of relevant hits in structure database is 0. The prediction was made based on sequence conservation only.
The PANTHER program also calculates sequence homology. The likelihood of the transition of one amino acid to another, the subPSEC from R199 to C199, is -
4.36316. P deleterious
, the probability that a given variant will cause a deleterious effect on protein function, is 0.79627. Finally, align-GVGD use Grantham matrix
scoring system to calculate the chemical differences between a given amino acid pair. Grantham Variation (GV) measures the degree of biochemical variation among amino acids found at a given position in the MSA1. Grantham Deviation
(GD) measures the ‘biochemical distance’ of the mutant amino acid from the observed amino acid at a particular position. GV of Human RNF170 protein R199 in MSA is 0.00, and GD of R199C is 179.53 (>0). According to the criteria of
Align-GVGD, the mutation is predicted to be deleterious when GV=0 and GD>0.
Additionally, the mutation R199C is classified to C65 class according to GD score, a mutation class which is most likely to interfere with protein function.
Multiple sequence alignment and sequence conservation
Multiple sequence alignment for vertebrate orthologs (MSA1) is shown in
Supplemental figure 3. The multiple sequence alignment shows that R199 is a highly conserved residue in vertebrates. The multiple sequence alignment for the selected Eukaryota orthologs (MSA2) is given in Supplemental figure 4. Proteins in MSA2 include more distantly related species, and the residue at R199 position is less conserved than it is in vertebrate orthologs.
Summary of protein motifs
The protein product of the refseq of the RNF170 gene has 258 amino acids including a RING finger motif at residue 87-130. The conservation of the RING finger motif is shown in the sequence logo (Supplemental figure 5). According to
UniProt annotation ( http://ca.expasy.org/cgi-bin/niceprot.pl?Q96K19
), RNF170
also has three potential transmembrane regions at amino acids 25-45, 202-222, and 224-244. The mutation R199C is close to the second transmembrane segment. As shown in supplemental figure 3, R199 is not very highly conserved in more distant orthologs. Other amino acids aligned to R199 include glutamine, lysine, and valine; however, cysteine is never observed at the position. The three-dimensional structure of the RNF170 protein has not been identified.
ModBase (http://modbase.compbio.ucsf.edu) contains a protein model of the ring finger domain that is present in 47 residues in RNF170 (C87-V133); however, this region is insufficiently large to determine the structural effect that the R199C mutation may have on the protein.
Supplemental Figures
Supplemental figure 1. RNF170 immunohistochemistry in patient and control spinal cord samples. A , Patient mid-cervical spinal cord. B , Patient lumbar spinal cord. C , Control upper thoracic spinal cord. D , Control lumbar spinal cord.
Immunodetections were made using an anti-RNF170 antibody in top panels; lower panels show negative control detections made using only anti-rabbit secondary antibody.
Supplemental figure 2. RNF170 protein levels do not differ between patient and control lymphoblasts. Detection was made using anti-RNF170 (Abcam). The expected molecular weight is 29.8 kDa. A , Soluble fraction of MG132 treated lymphoblasts. B , Insoluble fraction of MG132 treated lymphoblasts. C , Soluble fraction of untreated lymphoblasts. D , Insoluble fraction of untreated lymphoblasts. E , MG132 treated lymphoblasts. F , Untreated lymphoblasts.
Bottom panels marked by an asterisk represent actin loading controls.
* 20 * 40 * 60 * 80
Homo_sapiens M AKYQGEVHSLKLDDD SVIEGVSDQVLV A VVVS FAL IA T LVY A L F RN VH QNIHPENQE L VR V LREQLQ T EQ DAPAAT R Q QF
Bos_taurus
Rattus_norvegicus
M AKYQGEVQSLKLDDD SVIEGVSDQVLV A VVVS LAL IA T LVY A L F RN AH QNIHPENQE L VR V LREQLQ T EQ DAPAAA R Q QF
M ARYYSEVQSLQ-QDD SVIEGVSDQVLV A VVVS FAL IA S LLY A L L RN VQ QNIHPENQE L VR V LREQLQ T EQ DVPAPA R Q QF
Mus_musculus
Gallus_gallus
M ARYYSEGQSLQ-QDD S F IEGVSDQVLV A VVVS FAL IA T LLY A L L RN VQ QNIHPENQE L VR V LREQ F Q T EQ DVPAPA R Q QF
M DSHQAEVQSLKLDND SVIEGISDQVLV A VVLS FTF IA A LIY T L L RN EH QNIHPENQE L VR A LRQQLQ T EQ DASTGD R HR F
Xenopus_tropicalis M ADNQEGRPYFPLDEG SIIEGVSDQVIV V VLLS FVA V GS LLY L L L RN DE QNIHPENQ DR VR A VREQLQ N EQ ETPPPP R P QF
Danio_rerio GSVCVDGAAAPAPDEA SLIEGVSN A VLL V LVLS VTL LA G L TTL L C R SEQ Q R IHPE S QE R VR V VREQLQ A EQ -VSSES R H QF m S IEG6S1qV66 666S 6a L y L Rn QnIHPEnQe VR 6R2QlQ EQ R qF
Homo_sapiens
* 100 * 120 * 140 * 160
YT D M Y CPICL H QAS F PVETNCGHLFCG A CIIAYWRYGSWLGAI S CPICRQTVTLL LT VF GEDD Q SQDVLR L HQ DI N DYNRR
Bos_taurus YT D M Y CPICL H QAS L PVETNCGHLFCGTCIVAYWRYGSWLGAI S CPICRQTVTLL LP VF GEND Q SQDVVS L HQ DI S DYNRR
Rattus_norvegicus
Mus_musculus
YT E M Y CPICL H QAS F PVETNCGHLFCGSCIIAYWRYGSWLGAI S CPICRQTVTLL LT VF GEDD Q SQDVVR L RQ DV N DYNRR
YT E M Y CPICL H QAS F PVETNCGHLFCGSCIIAYWRYGSWLGAI S CPICRQTVTLL LT VF GEDD Q SQDVIR L RQ DV N DYNRR
Gallus_gallus YT D M S CPVCL Q QAT F PIETNCGHLFCGSCIIAYWRYGSWLGAI R CPICRQTVTL FLP LF SEDQ Q G--ATQ V LQ DV N DYNRR
Xenopus_tropicalis
Danio_rerio
YS D M T CPVCL Q QAT F PVETNCGHLFCGSCIIAYWRYGSWLGAI N CPICRQTVTLI FP LF QATE Q E-DSQN I LREAIG YNRR
YS D M S CPVCL Q QA VL PVETNCGHLFCGSCIIAYWRYGTWLGAI S CPICRQ M VTLL FP LF QDSE Q SAEPTL I LT DI S DYNRR
Y3 M CP6CL QA P6ETNCGHLFCG CI6AYWRYG3WLGAI CPICRQtVTL 6F Q 6 d dYNRR
* 180 * 200 * 220 * 240
Homo_sapiens FSGQPRSIM E RIMDLPTLLRHAFREMFSVGGLFWMFRIRIILCL M GA FF YLISPLDFVPEALFGILGFLDDFFVIFLLLIY
Bos_taurus
Rattus_norvegicus
FSGQPRSIM E RIMDLPTLLRHAFREMFSVGGLFWMFRIRIILCL M GA FF YLISPLDFVPEALFGILGFLDDFFVIFLLLIY
FSGQPRSIM E RIMDLPTLLRHAFREVFSVGGLFWMFRIRIMLCV M GA FF YLVSPLDFVPEALFGVLGFLDDFFVIFLLLIY
Mus_musculus FSGQPRSIM E RIMDLPTLLRHAFREVFSVGGLFWMFRIRIMLCL M GA FF YLISPLDFVPEALFGILGFLDDFFVIFLLLIY
Gallus_gallus
Xenopus_tropicalis
FSGQPRSIM E RIMDLPTLLRHAFREMFSVGGLFWMFRIRI F LCL F GA FL YL A SPLDFLPEALFGILGFLDDFFVIFLLLIY
FSGQPRSLM D RIMDLPTLLRHAFREMFSVGGLFWMFRIRIVLCL L GA LF YLVSPLD I IPEAVFGLLGFLDDFFVLFLLLIY
Danio_rerio FSGQPRSLL D RL R DVPTLLRHAFREMFSVGGLFWMFRVRILLCV C GA LA YLVSPLDFLPE G V L GLLGFLDDFFVI L LL F IY
FSGQPRS66 R6mD6PTLLRHAFRE6FSVGGLFWMFR6RI LC6 GA YL SPLDf6PEa6fG6LGFLDDFFV6fLLlIY
*
Homo_sapiens
Bos_taurus
ISIMYREVITQRL T R
ISIMYREVITQRL N R
Rattus_norvegicus ISIMYREVITQRL T R
Mus_musculus
Gallus_gallus
ISIMYREVITQRL T R
ISIMYREVVTQRL N R
Xenopus_tropicalis ISIMYREVVTQRL Y R
Danio_rerio ISIMYREVVTQRL AG
ISIMYREV6TQRL r
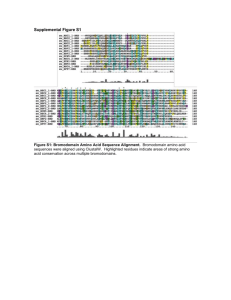
Supplemental figure 3. Multiple sequence alignment of orthologs selected from
Vertebrates. Amino acids are shaded according to sequence conservation.
Homosapien :
* 20 * 40 * 60 * 80 * 100
-----------------------------MAKYQGEVQSLKLDDDS V IEG V SDQVL----VAVVVSFALIATLVYALFRNVHQ--NIHPENQELVRVLRE : 65
Bostaurus :
Rattusnorv :
Musmusculu :
-----------------------------MAKYQGEVQSLKLDDDS
MQRYWRFQDTKIQDICFGALGELWIQRPVMARYYSEVQSLQ-QDDS
V
V
IEG
IEG
V
V
SDQVL----VAVVVSLALIATLVYALFRNAHQ--NIHPENQELVRVLRE
SDQVL----VAVVVSFALIASLLYALLRNVQQ--NIHPENQELVRVLRE
MQRYWRFQDNKIQDICFGVLGESWIQRPVMARYYSEGQSLQ-QDDSFIEG V SDQVL----VAVVVSFALIATLLYALLRNVQQ--NIHPENQELVRVLRE
Gallusgall :
Xenopus(Si :
Tetraodonn :
-----------------------------MDSHQAEVQSLKLDNDS
-----------------------------MADNQEGRPYFPLDEGS
V
I
IEG
IEG
I
V
SDQVL----VAVVLSFTFIAALIYTLLRNEHQ--NIHPENQELVRALRQ
SDQVI----VVVLLSFVAVGSLLYLLLRNDEQ--NIHPENQDRVRAVRE
------------------------------MEDGHCGDYLIQDEDT L IEG V SNQVL----FVVVVSVTFLAGLLTLLCRQEEQ--NIHPENQEHVRVVRQ
: 65
: 93
: 93
: 65
: 65
: 64
Daniorerio :
Strongyloc :
Nematostel :
---------------------------MEGSVCVDGAAAPAPDEAS
--------------------------------------MSQPERGT
L
I
IEG
VEG
V
I
SNAVL----LVLVLSVTLLAGLTTLLCRSEQQ--RIHPESQERVRVVRE
GDEFFQILGLIIVVAVPFLVAYRNRLRSIATG--AIHPESEAHVQHTRT
-----------------------------MASNVYSIFSLHHARGS I IEG I GDEVL----LALVTTLILIA-IVSLIYNSHFRMLNIHPLQAEQVRLARD
: 67
: 60
: 66
Brugiamala :
Caenorhabd :
---------------------------------------------M V HTN V AEIQF------------IEDSAITEFRRTFSG--AAQAEGPE-------
------------------MEQGVPATSSEIPPFTENITIPEVVSET V TEPDEDWLWPSDPDVELATQITMAIAVIFIVKAIFD--AWQSRRRQRAASRMD
: 34
: 80
Oryzasativ :
Arabidopsi :
Schistosom :
----------------------------------------------------------------------------------------------------
----------------------------------------------------------------------------------------------------
-------------------------------------------MGV L FES L DDHVVIFMSLPAIVLFLVGVRVVWGYFQSCSR--RIH------------
Monosigabr : ---------------------------------------------M MDG I SGVV-----ALLFMGIAVVWMAVFGWCAYKIS--SIWSRQPPSM-----
: -
: -
: 43
: 42
Homosapien :
Bostaurus :
* 120 * 140 * 160 * 180 * 200
QL-------------------------QTEQDAPAATRQ----QFYT-DMY CPICL HQASFPVE TNCGH L FC GA------------------------C
QL-------------------------QTEQDAPAAARQ----QFYT-DMY CPICL HQASLPVE TNCGH L FC GT------------------------C
: 110
: 110
Rattusnorv :
Musmusculu :
QL-------------------------QTEQDVPAPARQ----QFYT-EMY CPICL HQASFPVE TNCGH L FC GS------------------------C
QF-------------------------QTEQDVPAPARQ----QFYT-EMY CPICL HQASFPVE TNCGH L FC GS------------------------C
: 138
: 138
Gallusgall :
Xenopus(Si :
Tetraodonn :
QL-------------------------QTEQDASTGDRH----RFYT-DMS CPVCL QQATFPIE TNCGH L FC GS------------------------C
QL-------------------------QNEQETPPPPRP----QFYS-DMT CPVCL QQATFPVE TNCGH L FC GS------------------------C
QL-------------------------QTDQVPGPQDRQ----QFYS-DMS CPVCL QQAVLPVE TNCGH L FC GEWHTRATLTSSWLLFTCTFLLWCTGS C
: 110
: 110
: 134
Daniorerio :
Strongyloc :
Nematostel :
QL-------------------------QAEQ-VSSESRH----QFYS-DMS CPVCL QQAVLPVE TNCGH L FC GS------------------------C
QLRYGRPNADDHAASANGQATGESNGQQNGEGQSSDGRHGMSAQPYNGDRP CPICL DEKECAAE TNCGH V FC GN------------------------C
WLGIGTGGT-------------EQDNNERDEIQAPEPPR----AFSN-DRQ CPVCI TDARFLTM TNCGH E FC AP------------------------C
: 111
: 135
: 123
Brugiamala :
Caenorhabd :
Oryzasativ :
---------------------------TGASGSRARTLR----RFGD-DHI CPIC FGQASFAVV TNCGH L FC CN------------------------C
E--------------------------NAERNQIITQRRISEALHQS-SHE CPICL ANASFPVL TDCGH I FC CE------------------------C
---------------------------MSSSSASAVPPE---------DDV C S VC HDRFRIPCQA NC S H W FC GE------------------------C
: 77
: 128
: 39
Arabidopsi :
Schistosom :
Monosigabr :
---------------------------------MNAPPE---------NEV C S IC HGHFNAPCQ SNC S H W FC GN------------------------C
---------------------------SNQMSERIRDRS------NS-DYD CPICM EFPSLMVE TNCGH R FC AE------------------------C
---------------------------ATATGGPEQPPS----QRGD-HTT C A ICL DAPTNPII TNCGH C YC GM----------------SCLDEDRDW C
: 33
: 84
: 94
Cp6C 1CgH 5C C
Homosapien :
Bostaurus :
* 220 * 240 * 260 * 280 * 300
II AY WR Y-GSW L GAIS CPICR QT--------V T L L LTVFGE-------DDQSQDVLR L HQD I ND YNRRFSG QP R S IM ER I M DLP T LLR HAF R E M FSVG
IV AY WR Y-GSW L GAIS CPICR QT--------V T L L LPVFGE-------NDQSQDVVS L HQD I SD YNRRFSG QP R S IM ER I M DLP T LLR HAF R E M FSVG
: 192
: 192
Rattusnorv :
Musmusculu :
Gallusgall :
II AY WR Y-GSW L GAIS CPICR QT--------V T L L LTVFGE-------DDQSQDVVR L RQD V ND YNRRFSG QP R S IM ER I M DLP T LLR HAF R E V FSVG
II AY WR Y-GSW L GAIS CPICR QT--------V T L L LTVFGE-------DDQSQDVIR L RQD V ND YNRRFSG QP R S IM ER I M DLP T LLR HAF R E V FSVG
II AY WR Y-GSW L GAIR CPICR QT--------V T L -FLPLF---------SEDQQGATQ V LQD V ND YNRRFSG QP R S IM ER I M DLP T LLR HAF R E M FSVG
: 220
: 220
: 190
Xenopus(Si :
Tetraodonn :
II AY WR Y-GSW L GAIN CPICR QT--------V T L I FPLFQ--------ATEQEDSQN I LREAIG YNRRFSG QP R S LM DR I M DLP T LLR HAF R E M FSVG
II AY WR Y-GTW L GAIN CPICR QM--------V R L V L FPAGPV-------QDGEAEPQL I LRD I ND YNRRFSG QP R S LM DR L R DVP T LLR HAF R E M FSVG
: 191
: 217
Daniorerio :
Strongyloc :
Nematostel :
II AY WR Y-GTW L GAIS CPICR QM--------V T L L FPLFQDSEQSAVAADSPVEPTL I LTD I SD YNRRFSG QP R S LL DR L R DVP T LLR HAF R E M FSVG
LI AY WR H-GTW L GAIS CPVCR QM--------V T I I LPVFQED------EQNSGEGGR I MAE I RD YNRRFSG EP R PF M DY I Y DLP T LLR HTA R DFFSLH
II TY WR H-GRW L GAVQ CPVCR QQ--------V N L L FANFSSE------ESSSDDSHQWRGE I NE YNRRFSG LP R S VM EH L R DLP T LLR QLFSE L FSVG
: 200
: 218
: 206
Brugiamala :
Caenorhabd :
Oryzasativ :
I YGY W QYSASL I TPVK C A VCR EIVRFGSFTYA V N L L IPLPVEGER----ENSADEALRCDEQ L TD YNRRFS SER R P II DY I R DLP V LV PHMF R A V VSVN
II QY W QQSKAI V TPCD C A MCR ST---------FY M L LPVHWPTMGTS--EETDDHIQENNIR I DD YNRRFS -IN R P VL DY I R DIP I LI PYLI R NFFNND
II RV W NH-GPS V QPCK CPICR RL--------I N L L VPANVSID-----NDDDPQIQH V LGE V QH YNR I F G G TP R N L TQR L Q DLP FF IR RLF R E L LDPQ
: 172
: 215
: 123
Arabidopsi :
Schistosom :
Monosigabr :
IM LV WR H-GST L RPCK CPLCR RP--------I S L L VPSEETVR-----SRNDATVSE V LHD V ET YNR V F G G QSSG LI QR M Q DLP F LLR RLL R E M MDPQ
F I LH WK R-TVYSRIIS CPMCR GQ--------V STL TELFTAEEL----RDTSNRRSK I EAD L RL FNR WH SG GPIS II DR I R DIP LF V -YGFIQ L LLSG
FASLM R Q-SDFGTHRA CP T CR RR--------V NFL F------------SQQPLGDDA I SRE V RA FNRRY GPER R S M SEV V A D A P E LLR QFGAS L FDPT
: 117
: 168
: 171
w Cp CR 5NRr sg r 6 D P l6r r
Homosapien :
Bostaurus :
* 320 * 340 * 360 * 380 * 400
-G L FW M F RIRI I L CLMGAFF YLISP L D F VPE A L F GILG -------------------------------F LDD FF V IF LL L I Y I SI MYR E VI T-QRL---
-G L FW M F RIRI I L CLMGAFF YLISP L D F VPE A L F GILG -------------------------------F LDD FF V IF LL L I Y I SI MYR E VI T-QRL---
: 256
: 256
Rattusnorv :
Musmusculu :
Gallusgall :
-G L FW M F RIRI M L CVMGAFF YLVSP L D F VPE A L F GVLG -------------------------------F LDD FF V IF LL L I Y I SI MYR E VI T-QRL---
-G L FW M F RIRI M L CLMGAFF YLISP L D F VPE A L F GILG -------------------------------F LDD FF V IF LL L I Y I SI MYR E VI T-QRL---
-G L FW M F RIRI F L CLFGAFL YL A SP L D F LPE A L F GILG -------------------------------F LDD FF V IF LL L I Y I SI MYR E VV T-QRL---
: 284
: 284
: 254
Xenopus(Si :
Tetraodonn :
Daniorerio :
-G L FW M F RIRI V L CLLGALF YLVSP L D I IPE A V F GLLG -------------------------------F LDD FF V LF LL L I Y I SI MYR E VV T-QRL---
-G L FW M F RIRI L L CLVGAIT YL A SP L D I LPE A L F GLLG -------------------------------F LDD FF V IL LL L V Y I SI MYR E VV T-QRL---
-G L FW M F RVRI L L CVCGALA YLVSP L D F LPE G V L GLLG -------------------------------F LDD FF V IL LL F I Y I SI MYR E VV T-QRL---
: 255
: 281
: 264
Strongyloc :
Nematostel :
Brugiamala :
-G I VW M F RLRI IFCFAAALL YLISP L D I IPE A V F G FF G -------------------------------FF DD IF V VL IL A I Y V TG IYR G IV A-QRM---
-G L VW V L RMRI I L CFFAAAL Y F ISP L D I IPE S V F GILG -------------------------------L LDD AL I LL LL L V Y V TEA YR QY V A-NMA---
-G L MF M F RIR FF L CLCGMAV YILSP F D I LPE AAF GVLG -------------------------------M VDD IF I AF VV L V YATI LFR Q ML AGGRLHFA
: 282
: 270
: 240
Caenorhabd :
Oryzasativ :
-IFTL V YQ VRI GFVFICVIT Y F L L P S D M VPE S I Y GIIG -------------------------------F LDD CI I GI LV FGA M FRWL R EY M ADRGL---
RT L PL V F R A RM V M MVALSAI YVLSP I D I LPE N V L GL F G -------------------------------FF DD FL V LL IV F L H L AA VYR S LL L-YRH---
: 280
: 188
Arabidopsi :
Schistosom :
Monosigabr :
RT L PL V I R A RV Y I ALILSAI YIISP I D I IPE G V L GVIG -------------------------------L LDD LL I AL I CF L H V AA LYR S VL Y-SRH---
EGTFA L MQ IRL T L LGLCVLF YLITP F D F IP DV V A G F LG -------------------------------I LDD II V LC I FT I H V VS VY QT I SSRREL---
-N V SM L V KLRV I L SFVLAAL Y A I M P F D V LPE A V L GI F G CVCTLLTLVAERGHACLASMVNPNLSMVVSRLA DD AL V FL VV A V TSANTI R QNFV-QAS---
: 182
: 234
: 266
6 R Y P D 6Pe G G DD 6 6 r
*
Homosapien :
Bostaurus :
--------TR------
--------NR------
: 258
: 258
Rattusnorv :
Musmusculu :
Gallusgall :
Xenopus(Si :
Tetraodonn :
Daniorerio :
--------TR------
--------TR------
--------NR------
--------YR------
--------N-------
--------AG------
: 286
: 286
: 256
: 257
: 282
: 266
Strongyloc :
Nematostel :
Brugiamala :
--------E-------
--------TGIKQKEL
GAAGGRNSGRERERRE
: 283
: 278
: 256
Caenorhabd :
Oryzasativ :
--------ARN-----
--------GGH-----
: 283
: 191
Arabidopsi :
Schistosom :
Monosigabr :
--------GGS-----
--------TR------
--------QRAN----
: 185
: 236
: 270
Supplemental figure 4. Multiple sequence alignment of RNF170 orthologs selected from Eukaryota. Amino acids are shaded according to sequence conservation.
Supplemental figure 5. Sequence logo of RNF170 orthologs selected from
Eukaryota. The sequence logo was built with MSA2.
Supplemental Tables
Supplemental table 1. Complete set of genes sequenced for ADSA.
DKK4
DUSP26
EFCAB1
ERLIN2
FKSG2
FNTA
FUT10
GINS4
GOLGA7
GOT1L1
GPR124
HGSNAT
HOOK3
HTRA4
IDO
IKBKB
KCNU1
KIAA0146
ADAM18
ADAM2
ADAM32
ADAM5P
ADAM9
ADRB3
AGPAT6
ANK1
AP3M2
ASH2L
BAG4
BRF2
CEBPD
CHRNA6
CHRNB3
DDHD2
RB1CC1
RNF170
SFRP1
SLC20A2
SNAI2
SNTG1
STAR
ST18
TACC1
THAP1 tMDC
UBE2V2
UNC5D
VDAC3
WHSC1L1
ZMAT4
ZNF703
LETM2
LSM1
MAK16
MCM4
MYST3
NKX6-3
NRG1
PCMTD1
PLAT
PLEKHA2
POLB
POTEA
PPAPDC1B
PRKDC
PXDNL
RAB11FIP1
RNF170x1F
RNF170x1R
RNF170x2F
RNF170x2R
RNF170x3F
RNF170x3R
RNF170x4F
RNF170x4R
RNF170x5F
RNF170x5R
RNF170x6F
RNF170x6R
RNF170x7F
RNF170x7R
Supplemental table 2. Primers and PCR conditions for RNF170 .
CCAGTTCGCTGCGTGTC
CTACCCCAAGAAGGCCAG
TTTAAAGAACTCCATCAGTCCC
ATGTTAGCATTCAAAGCCCC
CAGCCTGAAACGTACAGTCC
AATTTGAAGCTGGAAAGAAGC
TGATGAGGGTTACACACTGC
GAGAAAATTGGGAATCCAGC
TGAATCTTTCTGAGGATATGTGG
GGTGACATCAAAGAAGATTTGG
AGTTACTTACTCCCCTCCCC
TAGGCGTTAGCTACTGTGCC
AAATATGTTCAATTAATGTGTAACTCC
AACTTCAAACATGAAAATTCCAG
PCR was performed using 50ng DNA, 20 pmol of each primer, 10X buffer, 0.25 nM dNTPs and 0.15 ul of Taq (Qiagen). For exons two to seven, a denaturation step of 5 minutes was first performed at 94°C. Then a touchdown protocol was used which consisted of an initial cycle of 30 seconds denaturation at 94°C, 30 seconds annealing at 59°C and 45 seconds elongation at 72°C. This was followed by nine cycles in which the annealing temperature was decreased each time by 0.5°C. Then 25 cycles of 30 seconds denaturation at 94°C, 30 seconds annealing at 59°C and 45 seconds elongation at 72°C were run. A final extension at 72°C was performed for 7 minutes. For exon 1, the same touchdown proto col was used except the temperature was decreased from 65°C to 60°C, followed by cycles at 60°C. In addition, 1x Q solution (Qiagen) was incorporated in the PCR mix.
Supplemental References
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: A sequence logo generator. Genome Research 14: 1188-1190
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792-97
Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV (2006)
Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic
Acids Res 34(5): 1317-25
Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions.
Genome Res 11: 863-74
Ng PC, Henikoff S (2002) Accounting for human polymorphisms predicted to affect protein function. Genome Res 12: 436-46
Ng PC, Henikoff S (2003) SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 31: 3812-14
Sunyaev S, Ramensky V, Bork P (2000) Towards a structural basis of human nonsynonymous single nucleotide polymorphisms. Trends Genet 16: 98-200
Sunyaev S, Ramensky V, Koch I, LatheW3rd, Kondrashov AS, Bork P (2001)
Prediction of deleterious human alleles. Hum Mol Genet 10: 591-97
Ramensky V, Bork P, and Sunyaev S (2002) Human non-synonymous SNPs: server and survey. Nucleic Acids Res 30(17): 3894-900
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de
Silva D, Zharkikh A, Thomas A (2006) Comprehensive statistical study of 452
BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43(4): 295-305
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, et al (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13:
2129-41
Thomas PD, Kejariwal A (2004) Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: evolutionary evidence for differences in molecular effects. Proc Natl Acad Sci USA 101: 15398-403