Supplementary Methods (doc 89K)
advertisement

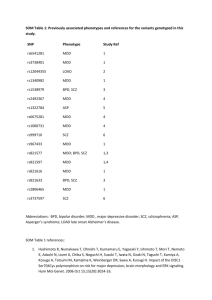
Gene Expression QC Quality control (QC) was conducted on NESDA subjects together with a previously reported twin sample from the Netherlands Twin Registry 1. The expression data were required to pass standard Affymetrix Expression Console quality metrics before further QC. All probe sequences were mapped to the human genome (hg19) using BOWTI2 , and probes not mapped uniquely or intersecting a polymorphic SNP (HapMap3 and 1000 Genomes Project) were removed 3,4. We mapped and annotated all Affymetrix U219 probesets to GENCODE gene models 5, probesets not matching any gene or transcript were removed, resulting in 45,574 probesets targeting 18,391 genes. Expression values were obtained using robust multichip averaging normalization (RMA, Affymetrix Power Tools, v1.12.0). Samples with sex inconsistency based on chrX and chrY probesets were removed. We verified sample identity between gene expression and SNP genotypes using 500 of the most significant local eQTL (SNP-transcript pairs) to estimate a post mismatch probability between gene expression and genotype profile. We examined the pairwise correlation matrix of expression profiles using a standardized median deviation correlations (D)1 in order to identify and remove array correlation outliers. The post mismatch probability and D were highly correlated with many expression probesets and were used as covariates in the final models. We evaluated the importance of multiple other potential covariates on gene expression, and chose 16 covariates for all analyses (sex, age, body mass index, smoking status, red blood cell count, laboratory, month and hour of blood extraction, time between blood extraction and RNA hybridization, hybridization plate and well, D, post mismatch probability, and 3 PCs derived from genotype data, Table S1). The covariates of greatest impact (>20,000 probesets) reflected batch effects, array processing, and RNA sampling, while subject characteristics (e.g., smoking, body mass index, age, or sex) influenced far fewer probes (<7,100). Linear models for MDD status and gene expression associations Primary inference was conducted on the basis of a linear model consisting of empirical covariates (Table S1), the appropriate MDD term as independent variables, and gene expression as dependent variable. P-values and betas were computed for the associations between MDD and gene expression for each of the 45,574 probesets. Two different linear models were considered: a model where the current MDD and remitted MDD groups were combined and a model using MDD as 3 level categorical variable (control, current MDD and remitted MDD) from which we derived overall MDD associations with gene expression (F-test) and three pairwise MDD group comparisons (controls vs. current MDD, controls vs. remitted MDD and remitted MDD vs. current MDD using ttests). When comparing the mean expression between the three MDD groups, gene expression was residualized using the linear model described above (without MDD). To verify confounding effects of immune status and antidepressants use, CRP or antidepressant use (SSRI, TCA and SNRI) were added to the linear model. P-values and betas for the current MDD-control group comparisons were recomputed after removal of the antidepressant users from the linear model. Control for multiple comparisons was addressed using the false discovery rate 6 , applying a threshold of FDR<0.1. In addition, the P-values and directions of effects for associations between MDD status and gene expression from a recent RNA-seq study (436 cases and 459 controls) 7 were meta-analysed with our findings using a weighted Z-score method. Genome-wide SNP assays used for eQTL analysis DNA was extracted from whole blood and all samples were randomized to plates. Genotyping was conducted using Affymetrix Genome-Wide Human SNP Array 6.0 per manufacturer protocol as described previously 1. Subjects were removed for Affymetrix contrast QC ≤ 0.4, high missingness (> 0.05), outlying genome-wide homozygosity, outlying ancestry via principal components analysis 8 , or discrepant sex. SNP QC included removal of SNPs for non-unique probe mapping to NCBI Build 37/UCSC hg19, low minor allele frequency (< 0.005), substantial deviation from HapMap3 CEU founder allele frequencies, deviation from Hardy-Weinberg equilibrium (PHWE < 1x10-8), or high missingness (> 0.05). Examine gene expression as mediator of DNA associations with MDD status Gene-based analysis of the PGC MDD GWAS results9 was conducted using VEGAS 10 and JAG 11. For JAG, a test statistic for each gene was computed using the sum of the -log10 P-values of the SNPs in the gene. Empirical P-values of these test statistics were computed using 1,000,000 permutations. For VEGAS, for genes with expression differences associated with current MDD, we selected SNPs and corresponding P-values from the PGC MDD results and used these as input to VEGAS (URLs). The gene-based statistic used in VEGAS is the sum of the P-values from the SNPs within the gene converted into uppertail chi-squared statistics. Monte Carlo simulations generate chi-squared statistics with the same correlation structure as the observed statistics, induced by the LD structure of SNPs in the gene. The empirical gene-based P-value is the proportion of simulated test statistics that exceed the observed gene-based test statistic. 1 Wright FA, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K et al. Heritability and genomics of gene expression in peripheral blood. Nat Genet 2014; 46: 430–437. 2 Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25. 3 A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061– 1073. 4 International HapMap 3 Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58. 5 Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22: 1760–1774. 6 Yekutieli D, Benjamini Y. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Stat Plan Inference 1999; 82: 171–196. 7 Mostafavi S, Battle A, Zhu X, Potash JB, Weissman MM, Shi J et al. Type I interferon signaling genes in recurrent major depression: increased expression detected by whole-blood RNA sequencing. Mol Psychiatry 2014; 19: 1267–1274. 8 Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909. 9 Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013; 18: 497–511. 10 Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM et al. A Versatile GeneBased Test for Genome-wide Association Studies. Am J Hum Genet 2010; 87: 139–145. 11 Lips ES, Cornelisse LN, Toonen RF, Min JL, Hultman CM, International Schizophrenia Consortium et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry 2012; 17: 996–1006.