Searching for fossil genes
advertisement

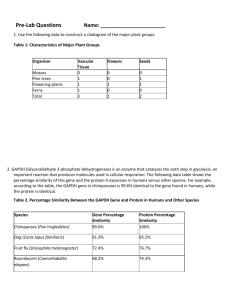
Searching for Fossil Genes A BioinFORMatics Activity By David Form Nashoba Regional High School dform@nrsd.net NAME: ______________________________ DATE: _____________ Introduction: You are the manager of a new animal food supply company. You need to find out if vitamin C needs to be included in new animal foods designed for dogs, cows, cats, mice and guinea pigs. Based on your research on the GULO gene, you will be able to determine if you need to provide vitamin C in these foods. Most mammals, such as mice, can produce their own vitamin C and therefore do not need a source of vitamin C in their diet. The GULO gene codes for an enzyme, Lgulonolactone oxidase, involved in vitamin C synthesis. The GULO gene is present in mice and most other mammals, but is either missing, or is nonfunctional, in some mammals. These animals cannot make their own vitamin C and must have this vitamin present in their diet. We will see if various mammals, including the primates, such as humans and chimps, have a functional GULO gene. A functional gene is able to produce a functional protein, in this case the GULO enzyme. In some mammals, the GULO gene is an example of a pseudogene. Pseudogenes are vestigial genes. That is, they were once functional in an ancestral species, but since they were no longer needed they accumulated mutations until they became nonfunctional. In many cases they evolve to the point where a protein can no longer be produced at all. Pseudogenes represent molecular evidence for evolution. As fossils are the remains of extinct organisms, pseudogenes are the remains of extinct genes. Procedure: The U. S. Government maintains a set of databases called GenBank, which contains nucleotide and amino acid sequences for those genes and proteins whose sequences have been determined. For your research we will use a computer program called BLAST. BLAST is able to search the GenBank databases. If you input a nucleotide or amino acid sequence into BLAST, it will search for any known genes or proteins that are similar to the one that you entered. You will use BLAST to determine if the genomes of cows, pigs, humans and chimps contain functional GULO genes, or if they contain vestigial GULO pseudogenes, which do not result in production of the GULO enzyme. Part I. Is the GULO gene present in various mammals? 1. Go to SharePoint and open the text file, “GULO gene mouse”. a. Note: For those of you who are not registered at NRHS, you can obtain the sequences at nrhs.nrsd.net. Under the “Departments” menu at the top of the page, select “Science”. Under “Science Links” you will find links to text file, “Mouse GULO Gene Mouse”. 2. Copy the nucleotide sequence for the mouse GULO gene. 3. On the internet, go to www.ncbi.nlm.nih.gov. 4. From the menu at the top of the page, select BLAST. 5. From the BLAST menu, select “nucleotide BLAST”. It is under “Basic BLAST”. 6. Copy the mouse GULO sequence into the box under “Enter Query Sequence”. 7. Scroll down until you see “Database”. 8. Check the “Others” box. Change the database setting in the box to “nucleotide collection”. 9. In the “Organism” window, type in “cow”. When you see cow appear in the box, select it. 10. Scroll to the bottom and find the “BLAST” button. Above the “BLAST” button, you will see the “optimize for” box. Select optimize for “somewhat similar sequences (BLASTn)” 11. Scroll back down and hit the “BLAST” button. 12. When BLAST is done with its search, you can scroll down and see a colorized diagram indicating the degree of similarity of the BLAST hits to your mouse GULO nucleotide sequence. Red and pink/purple mean a good match, while green, blue and black indicate a poor match. If the colored line spans the entire length of the window, then the “hit” sequence matches the inquiry sequence along its entire length. We want to see a high quality match along a majority of the inquiry sequence. 13. Below the colorized diagram is a “hit list” of your results. This shows the quality of matching as an E-value. An E-value is the chance that the matchup may be due to a random matching of a sequence of bases. The smaller the E-value, the more confidence you can have in your matching. A good match should have a low Evalue (red or pink line) and an alignment along a large segment of the sequence. 14. Note your result in the chart below. 15. Now start again and do BLAST searches for pig (Sus scrufa), human (Homo sapiens) and chimpanzee (Pan troglodytes) GULO genes. Record your data in the chart. Mammal GULO genes Species Mouse (Mus musculus) Dog (Canis familiaris) Cat (Felis catus) Cow (Bos Taurus) Pig (Sus scrufa) Guinea Pig (Cavia porcellus) Chimpanzee (Pan troglodytes) Human (Homo sapiens) Present or Absent? Present % coverage E-value 100% .0 PART II. Does the human GULO gene produce a functional protein? We will now use protein BLAST to search for GULO proteins in cows, pigs, humans and chimpanzees. 1. Copy the mouse GULO protein from the GULO protein text file in SharePoint. a. Note: For those of you who are not registered at NRHS, you can obtain the sequences at nrhs.nrsd.net. Under the “Departments” menu at the top of the page, select “Science”. Under “Science Links” you will find links to text file, “GULO Protein Mouse”. 2. Go to www.ncbi.nlm.nih.gov. 3. Select BLAST from the menu at the top of the page. 4. From the BLAST menu, select “protein BLAST”. It is under “Basic BLAST”. 5. Copy the mouse GULO sequence into the box under “Enter Query Sequence”. 6. Scroll down until you see “Organism”. 7. In the “Organism” window, type in “cow”. When you see cow appear in the box, select it. 8. Scroll to the bottom and select the “BLAST” button. 9. When BLAST is done with its search, you can scroll down and see a chart of your results. Note your result in the chart below. 10. Now start again and do BLAST searches for pig (Sus scrufa), human (Homo sapiens) and the other GULO genes. 11. Look for proteins with the same name (, L-gulonolactone oxidase). If the GULO protein is not present, other, more distantly related proteins may come up. They will have a much lower score and a higher E-value. Note that the E-value represents the chance that the result is due a random matching of some amino acid sequences from both proteins. An E-value of 0 means a statistically perfect match. A good E-value should be much lower than e-4. 12. Record your data in the chart. Functional GULO Proteins Species Mouse (Mus musculus) Dog (Bos taurus) Pig (Sus scrufa) Cow (Bos Taurus) Guinea Pig (Cavia porcellus) Human (Homo sapiens) Present or Absent? Present E value 0.0 Critical Thinking Exercises 1. Why do you think that primates (monkeys, apes and humans) have lost the ability to produce vitamin C? (Hint: think about the diet of early primates). 2. Explain why the GULO gene in humans may be considered vestigial. 3. Explain how the presence of the GULO gene in humans provides evidence for evolution. 4. You decide to use BLASTn to search for the GULO gene of another organism, such as a koala. BLAST does not turn up a gene. Does this necessarily mean that the gene is not present in koalas? Explain 5. Your new pet food company is designing healthful foods for dogs, cats, pigs, cows, mice and guinea pigs. Use the results of your research to determine if you need to provide vitamin C in these foods. 6. According to BLAST, the GULO gene in guinea pigs looks pretty good. Why can’t guinea pigs produce a functional GULO enzyme? Note to teachers: BLASTn turns up a complete GULO gene for guinea pigs with a good evalue. It turns out that there is a “stop” mutation in the middle of the guinea pig coding sequence, which prevents guinea pigs from producing a functional GULO enzyme. You can demonstrate this to your students in two different ways. You can use PubMed to find a paper on the GULO gene in guinea pigs. You can also use BLAST: “align two sequences” under “Specialized BLAST” to find the stop mutation.