SUPPLEMENTARY MATERIAL SUPPLEMENTARY NOTES
advertisement
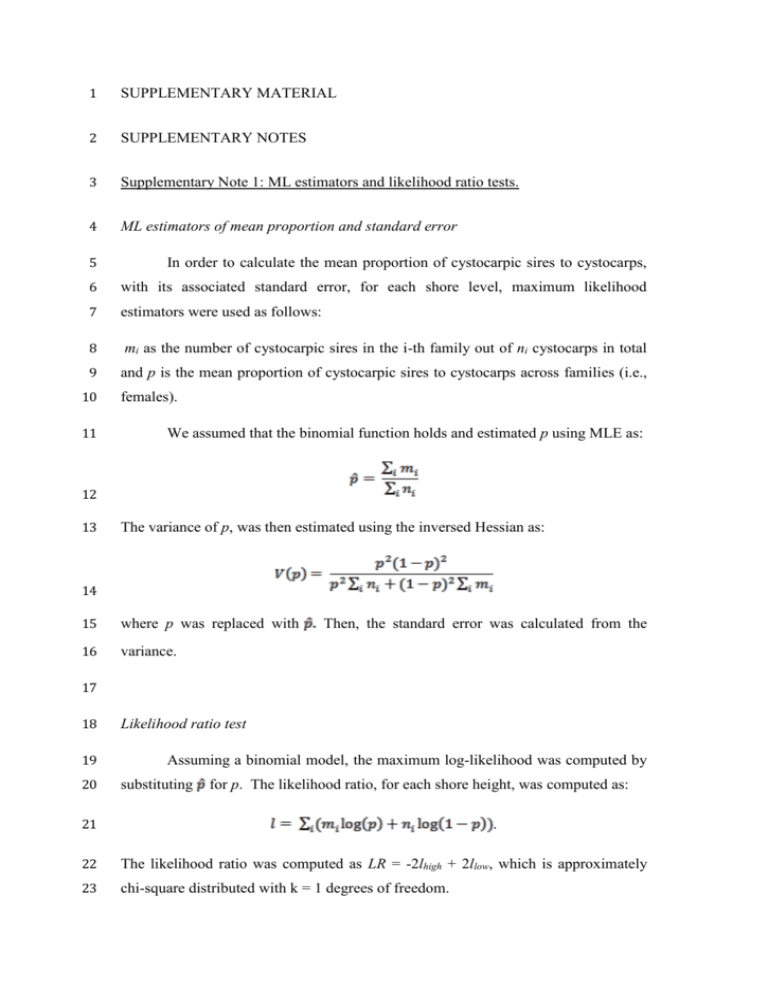
1 SUPPLEMENTARY MATERIAL 2 SUPPLEMENTARY NOTES 3 Supplementary Note 1: ML estimators and likelihood ratio tests. 4 ML estimators of mean proportion and standard error 5 In order to calculate the mean proportion of cystocarpic sires to cystocarps, 6 with its associated standard error, for each shore level, maximum likelihood 7 estimators were used as follows: 8 mi as the number of cystocarpic sires in the i-th family out of ni cystocarps in total 9 and p is the mean proportion of cystocarpic sires to cystocarps across families (i.e., 10 11 females). We assumed that the binomial function holds and estimated p using MLE as: 12 13 The variance of p, was then estimated using the inversed Hessian as: 14 15 where p was replaced with 16 variance. Then, the standard error was calculated from the 17 18 19 20 Likelihood ratio test Assuming a binomial model, the maximum log-likelihood was computed by substituting for p. The likelihood ratio, for each shore height, was computed as: 21 . 22 The likelihood ratio was computed as LR = -2lhigh + 2llow, which is approximately 23 chi-square distributed with k = 1 degrees of freedom. 24 Fitting a beta-binomial model 25 Young-Xu and Chan (2008) describe the beta-binomial model and its 26 application to the combination of binomial event rates across multiple studies as 27 ignoring overdispersion, or the presence of greater variability in a data set than 28 expected, when pooling data that are binomial in nature can lead to the erroneous 29 calculation of probabilities and confidence intervals. 30 overdispersion when pooling the 29 females used in this study, we followed the 31 method described by Roumagnac et al., (2004) and Sparks et al., (2008) using R, 32 ver. 3.03. (R Core Team 2014). For both the shore heights, there was unexplained 33 variation, possibly indicating overdispersion, or extra variation between females. 34 However, by comparing the binomial and beta-binomial models using a likelihood 35 ratio test as described above, there as no difference between the two models in the 36 high shore (χ2 = 0.180, df = 1, p > 0.860) or in the low shore (χ2 = 1.763, df = 1, p > 37 0.124). As the beta-binomial model did not provide a better fit for our data set, we 38 used the standard errors and maximum likelihood estimators assuming a binomial 39 distirubtion. In order to test for 40 41 REFERENCES 42 R Core Team (2014). R: A language and environment for statistical computing. R 43 Foundation for Statistical Computing, Vienna, Austria. URL http://www.R- 44 project.org/. 45 46 Roumagnac P, Pruvost O, Chiroleu F, Hughes G (2004). Spatial and temporal 47 analyses of bacterial blight of onion caused by Xanthomonas axonopodis pv. allii. 48 Phytopathology 94:138-146. 49 50 Sparks AH, Esker PD, Antony G, et al., (2008). Ecology and Epidemiology in R: 51 Spatial Analysis. The Plant Health Instructor. DOI:10.1094/PHI-A-2008-0129-03. 52 (http://www.apsnet.org/edcenter/advanced/topics/EcologyAndEpidemiologyInR/Spa 53 tialAnalysis/Pages/default.aspx) 54 55 Young-Xu Y, Chan KA (2008). Pooling overdispersed binomial data to estimate 56 event rate. BMC Medical Research Methodology 8:58. 57 58 59 60 61 62 63 64 65 66 Supplementary Note 2: Parentage assignments and indirect estimation of spermatial 67 dispersal in haploid-diploid red seaweeds. 68 Parentage analyses in haploid-diploid organisms do not share the same rich 69 literature and, consequently, multiple programs with which to perform parentage analyses as 70 is possible in diploid-dominant seed plants and animals. Red seaweeds, in particular, are 71 characterized by an alternation of free-living haploid and diploid stages, in which gametes 72 are produced mitotically by the gametophytes. In contrast, spores are produced meiotically 73 by the diploid tetrasporophytes. Thus, fertilization and meiosis are separated both in time 74 and space. 75 gametophytes. Moreover, with polymorphic markers, it is possible to identify the paternal 76 genotype without any ambiguity. 77 organism, such as mosses (see, for example, Szövényi et al., 2009). In contrast, diploid 78 male plants or animals contribute one, but not both alleles at any given locus. The zygote is an exact genetic copy of both its maternal and paternal This has also been shown in other haploid-diploid 79 Performing analyses matching genotypes, as we did in this manuscript using the 80 Multi-Locus Matches option as implemented in GenAlEx (Peakall and Smouse, 2006, 81 2012), is the most robust type of parentage assignment for red seaweeds, such as Chondrus 82 crispus. Applying existing programs, such as CERVUS (Kalinowski et al., 2007), would 83 result in uncertainty due to the violation of ploidy levels as well as assumptions one must 84 make at different stages of the analysis as implemented by certain programs. For example, 85 CERVUS requires diploid, codominant data. Simply artificially “diploidizing” haploids 86 would result in all female and male haploid genotypes turning into fixed homozygous 87 diploids. These individuals not only do not represent what is occurring on the shore in 88 natural populations, but also would clearly not be in Hardy-Weinberg Equilibrium. 89 CERVUS is one of the most popular programs for analyzing parentage. However, 90 there are certain assumptions made during simulations that do not fit our study system. The 91 program implements simulations of parentage analyses in order to assess the confidence of 92 real parentage assignments and are displayed as the natural log of the likelihood ratio. First, 93 we do not know the average number of candidate parents per offspring that were present. 94 As discussed in the main text of the manuscript, we sampled a 25 m2 area of the high shore 95 and low shore stand in which a frond was sampled every 25 cm, if present. As we sampled 96 only a single frond, we, thus, sampled only a single genet every 25 cm. As Chondrus 97 crispus forms a dense, almost mono-specific band within the intertidal zone and 98 morphological “holdfasts” can be mosaics of different genotypes and ploidies, we are not 99 currently able to estimate the average number of candidate parents per offspring from our 100 field observations. Indeed, this is one of the first detailed studies incorporating the male 101 component of C. crispus populations. Future studies will incorporate such data, where 102 possible, but understanding the average number of genets in this alga is much more difficult 103 than sampling a small forest of trees in which genets are discrete. Field ID of reproductive 104 state, let alone genets, is difficult to impossible without dislodging the frond. Finally, males 105 are likely reproductive weeks before cystocarps are sufficiently large enough to genotype 106 successfully. Our study is the only study, thus far, in this species addressing these questions 107 and will serve as a spring-board for future work and also how to sampled populations. 108 A second parameter calculated during the simulation of genotypes is the proportion 109 of loci mistyped. 110 genotype with a genotype selected at random under Hardy-Weinberg assumptions. Though 111 it is possible to simulate selfing and/or inbreeding, which is clearly occurring in Chondrus 112 crispus, the fixed homozygous state of each “diploidized” gametophyte would be in clear 113 violation of Hardy-Weinberg equilibrium. This raise the question of whether estimates of 114 error rate using these simulations would provide any realistic information to use in a 115 parentage assignment. 116 CERVUS defines a genotyping error as the replacement of a true Nevertheless, we artificially diploidized each haploid genotype into a fixed 117 homozygote. 118 number of offspring set to 10,000 (as recommended by the program), the number of We then performed simulations as implemented by CERVUS with the 119 candidate fathers to 557 (the total number of gametophytes sampled on the shore), the 120 proportion of candidate fathers sampled left to the default of 0.59, the proportion of loci 121 typed was set at 0.84 (this metric was calculated by CERVUS as we used the allele 122 frequency module to generate allele frequency data with our diploidized haploids added to 123 the cystocarpic data set) and the porportion of loci mistyped and error rate in likelihood 124 calculations set at 0.01. We, then, performed paternity analyses in which a male parent was 125 assigned to cystocarp by comparing the observed success of parentage assignment against 126 the success rate predicted by simulations. 127 Error rate analysis revealed estimated error rates of 0 across all loci, assuming all 128 known parent-offspring pairs are equally independent and detection probabilities ranging 129 from 0.72 to 0.93 (Table S2_1). 130 gametophyte C-8-122 in the high shore, as also found in our paternity analyses performed in 131 the main text of the manuscript. Four other putative sires were assigned paternity with 132 relaxed confidence (i.e., 20% false positive rate) in which there was mismatching at two 133 loci. Each putative sire was then checked for reproductive phenology and genotype in 134 comparison to each cystocarp sampled from each female. For the high shore female G-6- 135 306’s cystocarp “e,” a phenotypically male frond, A-10-62, was assigned relaxed paternity, 136 but mismatches occurred at two of the five loci. Similarly, the low shore female A-8-16’s 137 cystocarp “o” mismatched at two of the five loci with D-1-169, a phenotypically male frond. 138 Cystocarp “d” from the high female G-5-283 mismatched at one locus with B-3-90, a 139 vegetative frond. Finally, the low shore female A-9-18’s cystocarp “d” mismatched with a 140 low shore frond, A-9-61, at two loci out of four as this cystocarp was not genotyped at locus 141 Chc_04. First, A-9-61 was a reproductive female frond. Though, it is possible, due to high 142 inbreeding levels, that gametophytes could share the same genotype, the occurrence of 143 monoecious gamteophytes is uncertain, but also unlikely. There could, however, have been 144 a male gametophyte which shared the same MLG as A-9-61, but only 4 MLGs (each 145 repeated twice) were found out of a total sample of 586 gameotphytes sampled at the Port de Four cystocarps were assigned to the vegetative 146 Bloscon. Second, and most importantly, A-9-61 mismatched at two of four loci. Even if 147 there was an identical male MLG, it could not be the putative sire. Thus, using CERVUS 148 resulted in the same results found using the MultiLocus matches option in GenAlEx. 149 Despite some violations during simulations, we obtained the same results using both 150 approaches. 151 differences between parents and offspring, the MultiLocus matches option is more 152 appropriate accompanied by estimates of genotyping error (such as included Krueger- 153 Hadfield et al., 2013, see in particular the discussion on the use of TANDEM allelic 154 binning as well as Matschiner and Salzburger, 2009). However, for the present until a program can accommodate ploidy level 155 156 Table S2_1. Error rate analysis computed for the paternity analyses performed by CERVUS 157 for the sampled Chondrus crispus population sampled at the Port de Bloscon. Locus: locus 158 name; N compared: number of samples compared; N mismatching: the number of 159 individuals mismatching at each locus; Detection probability: the detection probability of 160 each locus and Error: the estimated error rate. 161 162 163 Locus N compared N mismatching Detection probability Error Chc_04 369 0 0.7164 0 Chc_23 455 0 0.9274 0 Chc_24 455 0 0.8320 0 Chc_35 455 0 0.7912 0 Chc_40 441 0 0.8733 0 164 Part II. Indirect estimates of dispersal using existing software. 165 Existing software, such as KINDIST/TWOGENER analyses as implemented in 166 POLDISP (Robledo-Arnuncio et al., 2007), require diploid, co-dominant data. The only 167 way with which to perform such analyses for a haloid-diploid species would be to substitute 168 a dummy allele for each haploid female gametophyte that was not carried by any of the 169 cystocarps. Though this might not bias the analyses, it does not reflect the true nature of the 170 intertidal haploid-diploid populations. Female and male gametophytes are haploid and 171 tetrasporophytes are diploid. Fitting haploid-diploid data into programs, that require co- 172 dominant diploid data, is akin to fitting a square peg into a round hole. 173 Nevertheless, we estimated the distribution of pollen dispersal distances using 174 KINDSIT, the first procedure of POLDISP, and by artificially placing a dummy allele for 175 each female at each locus in order to “create” diploid female gametophytes. We found no 176 decrease in among-sibship correlated paternity with distance (Figure S2_1). In other words, 177 the spermatial pool spatial genetic structure may have been too weak to enable reliable 178 estimation of a dispersal function. This was unsurprising considering we did not find 179 evidence of strong spatial structure using spatial autocorrelation analyses as implemented in 180 SPAGEDI (Hardy and Vekemans, 2002) in a previous study (Krueger-Hadfield et al., 181 2013). A future investigation sampling every frond from a given spatial scale (e.g., 10 cm2) 182 would enable the direct calculation of dispersal distances for a majority of sires based on the 183 levels of relatedness among females and cystocarpic sires. This would be the most accurate 184 metric as no programs currently exist to estimate dispersal curves in haploid-diploid 185 organisms. 186 187 188 189 Figure S2_1. A regression of among sib-ship correlated paternity, as calculated by 190 POLDISP (Robledo-Arnuncio et al., 2007), on the separation distance of the female 191 gametophytes. There was no decrease in among-sibship correlated paternity with distance 192 in a) the high shore (F1,34 = 1.31, p > 0.25) or b) the low shore (F1,169 = 0.42, p > 0.51). 193 a) 194 195 b) 196 197 REFERENCES 198 Hardy OJ, Vekemans X (2002). SPAGeDi: a versatile computer program to analyze 199 spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 200 618-620. 201 202 Kalinowski, ST, Taper, ML & Marshall, TC (2007). Revising how the computer 203 program CERVUS accommodates genotyping error increases success in paternity 204 assignment. Molecular Ecology 16: 1099-1006. 205 206 Krueger-Hadfield SA, Roze D, Mauger S, Valero M. (2013). Intergametophytic 207 sefling and microgeographic genetic structure shape populations of the intertidal red 208 seaweed Chondrus crispus (Rhodophyta). Mol. Ecol. 22: 3242-3260. 209 210 Matschiner, M. and W. Salzburger. 2009. TANDEM: integrating automated allele 211 binning into genetics and genomics workflows. Bioinformatics 25: 1982–1983. 212 213 Peakall R, Smouse PE (2006). 214 Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295. GENALEX 6.2: genetic analysis in Excel. 215 216 Peakall R, Smouse PE (2012). GenAlEx 6.5: genetic analysis in Excel. Population 217 genetic software for teaching and research-an update. Bioinformatics 28: 2537-2539. 218 219 Robledo-Arnuncio JJ, Austerlitz F, Smouse PE (2007). 220 package for indirect estimation of contemporary pollen dispersal. Mol Ecol Notes 7: 221 763-766. POLDISP: a software 222 223 Szövényi P, Ricca M, Shaw AJ (2009). 224 inbreeding depression in a dioicous moss species. Heredity 103 : 394-403. 225 226 227 228 229 Multiple paternity and sporophytic 230 Supplementary Note 3: Kinship pair comparisons using permutation tests. 231 Four different types of comparison were analyzed using permutation tests 232 conducted by randomly re-sampling kinship coefficients between each group using 233 R, ver. 3.0.3 (R Core Team 2014). Permutation tests were repeated 10,000 times 234 and an empirical type I error rate was calculated for each of the four different 235 comparisons using Bonferroni correction (Sokal and Rohlf 1995). 236 The first comparison was made in order to investigate whether the reference 237 allele frequency used led to different kinship coefficients between the three kinship 238 pairs. The Ritland (1996) and Loiselle et al., (1995) estimators were calculated 239 using the entire shore (high and low 5 m grids combined) or the 5 m grid within each 240 tidal height as reference allele frequencies. There were no significant differences 241 between the reference allele frequencies used for each shore height (Table S3_1). 242 Second, the Ritland and Loiselle estimators were compared for each kinship 243 pair in order to evaluate the differences, if any, between both estimators. The 244 Ritland estimator gives more weight to rare alleles in the population and may, 245 therefore, yield different kinship coefficients as compared to the Loiselle estimator. 246 However, there were no significant differences between the two estimators for each 247 of the kinship pair comparisons after Bonferroni correction (Table S3_2). 248 Third, comparisons were made between different kinship pairs within each 249 shore level stand in order to investigate levels of relatedness between kinship pairs. 250 Female-male and male-male comparisons were not significantly different, indicating 251 that the two kinship coefficients were similar magnitudes. For both female-male and 252 male-male comparisons to male-maleallpop, there were significant differences in 253 which females were more related to their cystocarpic sires and males were more 254 related to the females that they fertilized than to other males in the population (i.e., 255 males that fertilized carpogonia on other females; Table S3_3). 256 Finally, fourth, the levels of relatedness between each type of kinship pair 257 were contrasted between shore heights. 258 between the levels of relatedness between each kinship pair (Table S3_4). The 259 female-male and male-male comparisons were hypothesized to be more related to 260 each another than the same kinship pairs low on the shore. However, this was likely 261 due to the variable levels of relatedness found between female-male comparisons, 262 the sample disparity between shore heights (nhigh = 6, nlow = 16) and other 263 topographical factors influencing spermatial dispersal. 264 significantly higher levels of inbreeding in the high shore demonstrated by Krueger- 265 Hadfield et al., (2013), increased sampling accompanied with locating the 266 cystocarpic sires would likely reveal a pattern of increased relatedness with shore 267 height. This would likely be driven by the increased fragmentation of populations 268 with increasing tidal height and tidal induced gamete retention. 269 270 271 272 273 274 However, there were no differences However, based on the 275 Supplementary Note 3, Table 1. Comparisons between reference allele frequencies 276 for the Ritland and Loiselle estimators. The kinship pair comparison column lists 277 the two kinship pairs compared; the shore height column refers to the high or low 278 shore sampling location; the p-values for each comparison are listed under the 279 columns Ritland_shore vs. Ritland_5m and Loiselle_shore and Loiselle_5m. 280 “Shore” refers to the reference allele frequencies sampled from the high and low 281 shore 5 m grids and “5m” corresponds to the reference allele frequencies used for 282 the respective shore height (i.e., high shore 5 m grid for the high shore comparison). 283 The adjusted significance level following Bonferroni correction was 7.5 x 10-4. Kinship pair comparison 284 285 286 287 288 289 Shore height Ritland_shore vs. Ritland_5m Loiselle_shore vs. Loiselle_5m Female-male vs. Male-male High 0.3074 0.4281 Female-male vs. Male-male Low 0.3624 0.4790 Female-male vs. Male-maleallpop High 0.3083 0.4035 Female-male vs. Male-maleallpop Low 0.3473 0.4860 Male-male vs. Male-maleallpop High 0.0326 0.1008 Male-male vs. Male-maleallpop Low 0.2178 0.4268 290 Supplementary Note 3, Table 2. Comparisons between the Ritland and Loiselle 291 estimators. 292 compared; the shore height column refers to the high or low shore sampling 293 location; the p-values for each comparison are listed under the columns 294 Ritland_shore vs. Loiselle_shore and Ritland_5m vs. Loiselle_5m. “Shore” refers to 295 the reference allele frequencies sampled from the high and low shore 5 m grids and 296 “5m” corresponds to the reference allele frequencies used for the respective shore 297 height (i.e., high shore 5 m grid for the high shore comparison). The adjusted 298 significance level following Bonferroni correction was 7.5 x 10-4. The kinship pair comparison column lists the two kinship pairs Kinship pair comparison 299 300 301 302 303 304 Shore height Ritland_shore vs. Ritland_5m Loiselle_shore vs. Loiselle_5m Female-male vs. Male-male High 0.4904 0.3381 Female-male vs. Male-male Low 0.4398 0.2674 Female-male vs. Male-maleallpop High 0.3440 0.4195 Female-male vs. Male-maleallpop Low 0.2849 0.1269 Male-male vs. Male-maleallpop High 0.4651 0.0014 Male-male vs. Male-maleallpop Low 0.0974 0.0692 305 Supplementary Note 3, Table 3. Comparisons between the levels of relatedness 306 between kinship pairs using a) the Ritland_5m estimator and b) the Loiselle 307 estimator. 308 compared; the p-values for each comparison are listed under the columns high 309 (referring to the pairs compared within the high shore stand) and low (referring to 310 the pairs compared in the low shore stand). 311 following Bonferroni correction was 0.003. 312 a) The kinship pair comparison column lists the two kinship pairs Kinship pair comparison The adjusted significance level High Low 0.172 0.029 Female-male vs. Male-maleallpop < 0.001 < 0.001 Male-male vs. Male-maleallpop < 0.001 < 0.001 High Low Female-male vs. Male-male 0.1046 0.004 Female-male vs. Male-maleallpop < 0.001 < 0.001 Male-male vs. Male-maleallpop < 0.001 < 0.001 Female-male vs. Male-male 313 314 b) Kinship pair comparison 315 316 317 318 Supplementary Note 3, Table 4. Comparisons between the levels of relatedness of 319 the three kinship pairs between tidal heights using a) the Ritland_5m estimator and 320 b) the Loiselle_5m estimator. The kinship pair comparison column lists the kinship 321 pair; the p-values for each comparison are listed under the columns high vs. low. 322 The adjusted significance level following Bonferroni correction was 0.017. 323 a) Kinship pair comparison 324 High vs. low Female-male 0.3772 Male-male 0.4775 Male-maleallpop 0.2876 b) Kinship pair comparison High vs. low Female-male 0.4943 Male-male 0.4780 Male-maleallpop 0.4399 325 326 REFERENCES 327 Krueger-Hadfield SA, Roze D, Mauger S, Valero M (2013). Intergametophytic 328 sefling and microgeographic genetic structure shape populations of the intertidal red 329 seaweed Chondrus crispus (Rhodophyta). Mol. Ecol. 22: 3242-3260. 330 331 Loiselle BA, Sork VL, Nason J, Graham C (1995). Spatial genetic structure of a 332 tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot. 82: 1420– 333 1425. 334 335 R Core Team (2014). R: A language and environment for statistical computing. R 336 Foundation for Statistical Computing, Vienna, Austria. URL http://www.R- 337 project.org/. 338 339 Ritland K (1996). Estimators for pairwise relatedness and individual inbreeding 340 coefficients. Genet Res. 67: 175-185. 341 342 Sokal RR, Rohlf FJ (1995). Biometry: the principles and practice of statistics in 343 biological research. 3rd ed. W. H. Freeman and Co., New York. 344 345 346 347 348 349 350 351 SUPPLEMENTARY FIGURE LEGENDS 352 Figure 1. a) The location of the females from which cystocarps were genotyped in 353 order to determine paternity in the high and the low 5 m grids. Next to each female 354 code is the kinship coefficient, if calculated, of Loiselle et al., (1995) as calculated 355 using the allele frequencies from the respective shore height 5 m grids. The only 356 sire that was identified from sampled fronds was located 25 cm from the female that 357 it fertilized. An * denotes the male frond which fertilized the female located 25 cm 358 away, C-9-123. b) The location of the females within the 5 m x 5 m grids in relation 359 to all the other sampled fronds in both the high and low shore stands. 360 361 Figure 2. The line represents the number of unique genotypes over the total number 362 of genotypes for all the samples of tetrasporophytes and gametophytes from the 5 m 363 grids by sequentially adding each locus starting with the most polymorphic one (n = 364 8). Individuals missing data at one or more loci were removed prior to analysis. 365 The most polymorphic loci were Chc_04, Chc_23, Chc_24, Chc_35 and Chc_40 and 366 distinguished all individuals fronds. These five loci were used to genotype the 367 cystocarps. 368 369 370 371 372 373 FIGURES 374 Figure 1. 375 376 377 378 379 380 Figure 2.