Detection of mt polymorphisms for mt genotyping

Arabidopsis cytoplasmic diversity Moison et al
Detection of mt polymorphisms for mt genotyping
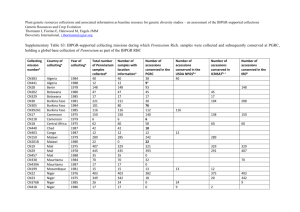
We searched for mt polymorphisms in a subset of 14 accessions (Additional Table 1). We first tested the few previously reported intraspecific polymorphisms in A. thaliana mt genomes
(Ullrich et al.
1997; Forner et al.
2005; Forner et al.
2008). The genomic organizations resulting from the recombination events I and II described by Ullrich et al.
(1997) could be easily checked by direct PCR amplification. Similarly, specific primer pairs were designed to check for the presence/absence of atp6-2 (Forner et al.
2008) or the atp9-cox3 organization described in L er
(Forner et al.
2005). Forner et al.
(2008) also pointed out a difference between Col-0 and C24 in the region upstream of ccmC. A ca 630 bp fragment in this region was amplified and sequenced for the 14 accessions of the sub-sample. Sequences were aligned with Genalys (Takahashi et al.
2003). The sequence of eleven accessions, including C24, differed from the reference sequence by only one base. The three remaining accessions, namely Col-0, Ler-1 and Ita-0 had very similar sequences, with only one or two differences, but they differed from the sequence of the other accessions by more than 25 positions. Among the SNPs that differentiated the ccmC sequences of
Col-0, Ler and Ita-0 from that of the other accessions, two modified a Cla I site that was subsequently used for PCR-RFLP. After PCR-RFLP typing of the entire sample of accessions, all ccmC amplification products that lacked a Cla I site were subsequently sequenced. Altogether, we identified five different sequences, differing by one or two SNPs, among the accessions that lack a Cla I site, raising to six the number of alleles in this region.
All ccmC sequences were deposited in the EMBL database (FN669556, FN669557, FN669558,
FN669559, FN669560, FN669561, FN669562, FN669563, FN669564, FN669565, FN669566,
FN669567, FN669568, FN669569, FN669570, FN669571, FN669572, FN669573, FN669574,
FN669575, FN669576, FN669577, FN669578, FN669579, FN669580, FN669581, FN669582,
FN669583, FN669584, FN669585, FN669586, FN669587, FN669588, FN669589, FN669590,
FN669591, FN669592, FN669593, FN669594, FN669595, FN669596, FN669597, FN669598).
Polymorphisms were also identified in two intergenic regions, nad9-rpl16 and atp8-orf107c, after sequencing the 14-accession subset following the same strategy as for the ccmC upstream region.
In both nad9-rpl16 and atp8-orf107c regions, two different sequences were identified in the 14accession subset; one corresponded to the reference sequence. The two sequences of the atp8orf107c intergenic region differ by a single SNP that modifies a Spe I site, which was subsequently used for genotyping the complete set of accessions by PCR-RFLP. The two
1
Arabidopsis cytoplasmic diversity Moison et al sequences of the nad9-rpl16 intergenic region differ by a 6 bp indel. This polymorphism could be genotyped by digestion of the amplification product with BstB 1 and Fok I, followed by 3% agarose gel electrophoresis.
New sequences of the atp8-orf107c intergenic regions (FN669554, FN669555) and of the nad9rpl16 intergenic region (FN669607) were deposited in the EMBL database.
To search for polymorphisms outside of mt gene coding regions, we used divergent primers, anchored in conserved coding sequences, on total DNA after Eco RI digestion and circularization, and then sequenced PCR products. This approach revealed a SNP that modifies an Eco RI site downstream of the cox2 gene that was subsequently genotyped by PCR RFLP.
Primer pairs were developed for direct amplification of all non-conserved orfs reported in the
C24 mt genome sequence (Unseld et al.
1997). Polymorphisms were observed among the subset of accessions for seven non-conserved orfs among the 80 tested. The complete set of accessions was therefore tested by PCR amplification for these seven orfs.
All mt polymorphisms are described in Additional Table 6.
2