Second meeting in Bruxelles
advertisement

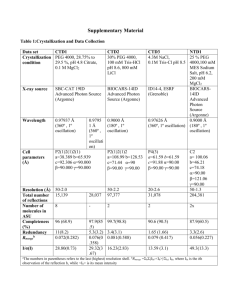
Second meeting in Bruxelles Consortium funded by the EU Under the contract QLG2-CT-2002-01298 The second meeting of the consortium Protein Folding Fragment hold in the Université Libre de Bruxelles, on behalf of Marianne Rooman, on November 24th and 25th 2003. 1. Presentation of the advancement of work during the last semester 1.1. Group 1 How to hopefully get automatic topohydrophobic positions from structures Jacques Chomilier, Anne Lopes The Paris team has moved during the past summer from Jussieu to Boucicaut, another campus in downtown Paris. From the common data bank of 116 PDB entries, about one half of the entries have been analysed in terms of topohydrophobic positions. Thus the second half remains to be determined. The major challenge is to find a way to have both fast access to these positions (which means as much as possible automatic) and secure (which in turn means slow and carefull check of the results). The bottleneck in this procedure is to be able to perform reliable structural alignment by blocks with allowed gaps. It seems from a glance at CKAAP, the Conserved Key Amino Acid Position server from Phil Bourne at San Diego Super Computing Centre (http://ckaaps.sdsc.edu/perl/browser.pl), that it is designed to produce the longest possible blocks of superposition, not to determine the largest amount of deep core positions. Thus we have been interested in an algorithm that is the baically the transposition to the 3D structures of the BLAST algorithm. It first tries to retrieve words of four to five amino acids, and from these seeds it then tries to extend them to form SHSSP, structural high scoring segment pairs. Then one can deduce a sequence alignment and further derive the highly conserved regions. This algorithm, called Yakusa, has been developped by Joël Pothier and al. It allows to treat a list of protein, thus one can fix an upper limit to the sequence identity (30% to be coherent with previous data) of any pair of entries. It is fairly fast and has the advantage to compare internal coordinates instead of the more common rmsd for comparison of structures, as we do agree with group 3 to think that rms is no good to find evolutionary relatedness. 1.2. Group 2 Calculations of Mostly Interacting Residues and comparison to topohydrophobic positions and Tightened End Fragments Nikolaos Papandreou, Elias Eliopoulos Group 2 is currently involved in WPs 1 and 2. Concerning WP1, the common protein dataset is complete and the server that will host the different aspects of the project is operational at the address http://biotech.aua.gr/LIFE/. Mirrors of the server should be shortly developed by a number of participants. Concerning WP2, the Monte-Carlo algorithm that is used to calculate the MIR (Mostly Interacting Residues) has been refined and a first set of definitive results obtained. They concern a subset of 43 proteins of the common dataset, for which both topohydrophobic residues and closed loops are known. The comparison between the MIR with topohydrophobic positions and loop ends confirmed the preliminary calculations and showed a very clear correspondence between them. Thus The MIR algorithm should be considered as a possible method to predict the critical residues that stabilize the protein hydrophobic nucleus. In the frame of WP1, the Monte-Carlo calculations on the rest of the dataset will be completed after the calculation of all topohydrophobic residues. Discussions with group 5 started in order to prepare the next WP in which groups 1, 2, 3 and 5 are involved. 1.3. Group 3 Closed loops of TIM barrel proteins Z Frenkel. In the abscence of Z. Frenkel, who could not get his passport/visa on time, these results have been presented by Edward Trifonov. In our effort to describe TIM barrel family by the closed loops of, presumably, limited number of types, we got across supervariability of the sequences belonging to apparently the same structural family of the loops. The similarity of the sequences within each family becomes obvious when instead of 20-letter amino-acid alphabet we used 2-letter alphabet, derived from the reconstruction of the origin and evolution of triplet code (Trifonov, Gene 261:139, 2000). The effect is so dramatic that for some of the closed loops of the same structure the 20-letter alphabet sequences are completely different (1-2 matches at most) while in the binary representation only 1-2 letters of 25-30 do not match. We suggest whenever the relatedness of structures is questionable sequence-wise, to use the binary code that would reveal the relatedness. Second observation is that some sequence-wise related segments (in 20-letter alphabet) turn out to be structurally very much different (by RMS). Yet, if the structures are presented by torsional angles (Angle walk) they turn out to be almost identical, with only 2-3 angles different, that completely distorts the closed loop appearance and RMS difference, but the rest of the angles are all the same for the two compared structures. Two letter alphabet for protein sequence comparisons Edward Trifonov The earlier published reconstruction of the origin and evolution of the triplet code (Trifonov, Gene 261, 139, 2000) suggests that the new codons appeared as point-mutated earlier codons, with conservation of purines and, respectively, pyrimidines in the central positions of the codons. This introduces two independent amino-acid alphabets: A, F, I, L, M, P, S, T, V – Ala-family, with pyrimidine-central codons, and C, D, E, G, H, K, N, Q, R, S, W, Y – Gly-family, with purine-central codons. It turns out, that, apparently, even after the triplet code was completed, the conservation of the two alphabets is still in place. Indeed, the tabulated replacements (PAM- and BLOSSUM-matrices show very strong separation of the replacements in the Ala-Ala-type and Gly-Gly-type replacements. The observed separation of the amino acids in two alphabets is of fundamental value both for protein evolution studies and for practicalities of sequence alignments. 1.4. Group 4 Progress with fingerprints Terri Attwood, Manuel Corpas The Manchester team has begun to make a systematic analysis of the common dataset of 116 structures provided earlier in the year. The dataset was divided first into 2 sets: those that already have some kind of fingerprint in the PRINTS database, and those that have not – the latter was made a priority for which to derive new fingerprints. The approach to producing new fingerprints is 2-fold – automatic and manual. The automatic approach will also be approached from 2 perspectives: (i) using the PDB domain as the seed for the fingerprint process, and (ii) using the equivalent full Swiss-Prot sequence as the seed for the fingerprint. Once new fingerprints have been created manually and automatically in this way, those fingerprints that are already in PRINTS in some form will be revisited. Here it is necessary to decide if the existing fingerprint falls entirely in the PDB domain, or if it contains motifs outside the domain. In the latter case, the fingerprint will need to be revised to better represent the structure. An overview of the process is shown in the Figure below. Fingerprinting Overview 117 PDB entries 56 61 Those in PRINTS Those NOT ***priority*** ~40 Auto 43 Match PDB domain Manual 13 ***priority*** Partial match PDB Swiss-Prot ***priority*** Objective Comparison RESULTS 117 Manual and < 117x2 Automatic Data integration & visualisation To date, we have done ~40 new fingerprints manually and have some kind of meaningful representation for 43; thus, we have completed almost half of the manual effort. During the coming months, we will concentrate on completing both the manual and automatic efforts. For the future, we are working with Steve Pettifer (Dept. of Computer Science) to determine how best to store our results and integrate them with those of the other teams. Dr.Pettifer is an expert on data integration and visualisation, and will help us to extend our current integration/visualisation software (UTOPIA, of which the CINEMA alignment editor is a core component), to handle the data emerging from each of the teams, as illustrated below. Using CINEMA & UTOPIA as a framework for visualising fingerprints, MIRs, LIRs, TEFs, Ts, etc., in 2- & 3D 1 2 3 4 1.5. Group 5 Prelude, Fugue and PoPMuSiC: are they in harmony with other methods? Dimitri Gilis, Marianne Rooman, Jean-Marc Kwasigroch, Yves Dehouck, Christophe Biot, René Wintjens The first 6 months of this project have been dedicated to: (1) the energy functions used to evaluate the compatibility between a sequence and a conformation, and (2) the comparison between the results obtained with our programs on the common data bank of 116 PDB entries, and the topohydrophobic positions and the limits of the TEF's. The dependence of distance-dependent database-derived potentials on the size of the proteins belonging to the database used to derive them is a drawback that has been identified some years ago and that is not fully understood yet. We have addressed this issue by probing the theoretical validity of these potentials as mean force potentials that take the solvent implicitly into account and involve entropic contributions due to atomic degrees of freedom and solvation. The results of this analysis have been used to devise new corrective functions that take into account the size of the protein studied. We have shown that these corrected potentials perform better than their more classical version to retrieve the correct sequencestructure association among a decoy set. We have also assembled a collection of decoy sets to evaluate the performance of energy functions used in the field of protein tertiary structure prediction. There exists a large number of decoy sets that are available on the web, but their quality is variable. In a first step, we have analyzed all these sets. We have then selected some of them in order to propose a collection of sets that are accurate, that have been created for proteins belonging to several structural classes and that contain non-native structures that are representative of the conformational space, with structures close to the native. The results of this analysis can be found at the URL: http://babylone.ulb.ac.be/decoys. Another part of our work has consisted of running Prelude, Fugue and PoPMuSiC on the 116 pdb's of our common data bank. We have identified with Fugue regions of the protein sequences that show a strong preference towards the native conformations, whereas the PoPMuSiC results give the positions along the sequence that are (not) optimized with respect to the thermodynamic stability of the protein. We have correlated these results with the topohydrophobic positions and the limits of the TEF's, for one protein. Our future work will consist of correlating systematically the results of Fugue and PoPMuSiC with the topohydrophobic positions and the TEF's. Moreover, we will compare the regions predicted by Fugue and the foldons derived from the 3D structures by the group of P. Wolynes. Finally, we plan to analyze, in collaboration with the Group 6, a large database of mutated proteins, for which the experimental folding free energy difference has been measured, with PoPMuSiC and FoldX. This collaboration has been initiated via a one month stay of D. Gilis in the laboratory of Group 6. 1.6. Group 6 TANGO : an algorithm to predict protein aggregation Frédéric Rousseau, Luis Serrano We have modified the FOLD-X algorithm so now it includes heteroatoms like Ca, Zn, Mg, Mn etc… with great accuracy, as well as the Kds for Ca ions. Also we have done a refinement of the force field and we are in the process of comparing our predictions of mutants with those of Partner: Rooman, with the idea of finding out complementarity and sinergisms. Regarding Protein Folding we have yet not been able to sort out the problem of doing a correct estimation of loop entropy in folding which be believe is necessary to accurate describe the folding pathways of proteins. In parallel we have developed independently of the proposal a software package called TANGO that predicts the tendency of a protein to aggregate. TANGO predicts with surprisingly good accuracy the regions experimentally described to be involved in the aggregation of 176 peptides of over 20 proteins. The predictive capacities of TANGO are further illustrated by two examples: the prediction of the aggregation propensities of A1-40 and A1-42 and in several disease-related mutations of the Alzheimer’s -peptide as well as the prediction of the aggregation profile of human acyl phosphatase. Thus, by capturing the energetics of structural parameters observed to contribute to protein aggregation and taking into account competing conformations, like -helix and turn formation, it is possible to identify with surprising accuracy protein regions susceptible of promoting protein aggregation. The success of TANGO shows that the underlying mechanism of cross- formation aggregates is universal. Logically this type of prediction is essential to understand protein folding, as well as for protein design, since it takes into account the effect of mutations on the denatured state of proteins. It is our intention to link TANGO to FOLD-X, so that when designing a protein or modifying a folding pathway by mutagenesis we could see the possible effect on the aggregation properties of the target molecule. For next year we plan to run FOLD-X on the PDB database generated by the consortium, producing an output that contains the energy per position, which could be compared to the results produce by other members of the consortium. Also we hope to finally have the loop problem solved and add to the new FOLD-X web server the possibility not only of predicting point mutations, but also folding pathways. 1.7. Group 7 Off Lattice molecular dynamics folding simulations David Perahia, Charles Robert, Liliane Mouawad We continued to develop the off-lattice molecular dynamics program (MMSIM) that was started at the beginning of our participation in the European consortium, by adding new modules allowing to increase its predictive power for finding the native state of a protein from the sole knowledge of its sequence. The protein is represented by a chain in which the residues are represented by single points located at the C positions. The force field contains secondary structure propensity potentials depending of the nature of residues within segments of 3 and 4 residues, and residue-residue contact potentials between the 20 types of residues that are used in a Lennard-Jones function. Our efforts were directed towards developing an optimisation procedure for energy parameters in such a way that the native state corresponds to the lowest energy in a set composed of decoy structures and the native one. A second condition was the introduction of a gap energy between the native structure and structures beyond 3Å root mean square deviation (rmsd) from the native structure. The optimization procedure consisted of generating decoy structures of a given protein by molecular dynamics simulation at various temperatures ranging from 10 to 1000K, starting from the native structure. Structures collected every 100ps from these trajectories were quenched by energy minimization. The optimized parameters are the energy-term weighting factors and the residue-residue contact energies. The optimization procedure was based on the minimization of an error rate function taking values from 0 to 1; the lowest value corresponds to the requirement that the native structure is energetically the most favoured structure. A Monte Carlo method was used to change the parameter values. An iterative scheme was designed consisting of successive generation of decoy sets with energy parameter optimization at each iteration step. The energy parameter optimization was carried out on a small 5-helix protein (1r69) in order to test the performance of our procedure and evaluate the limits of our potential energy function in discriminating the native structure. The first conclusions obtained were that secondary-structure propensity potentials discriminate helices well (all 5 helices are well predicted), but the residue-residue contact energies using only C atoms had no discriminating power in favoring native-like contacts between distant residues along the chain. Among 2000 structures obtained by molecular dynamics folding simulations with the optimized parameters, starting from an unfolded structure, only six displayed native-like structures. However, the introduction of an energy constraint favoring a TEF-like structure in the folding simulations increased the number of native-like structures appreciably. We now have the tools developed that permit us to find the optimal parameters of a given energy function and to evaluate its performance. Our next step is to consider a model with two points per residue, one corresponding to the C atom and the other to the sidechain, and to include a solvation energy term. We should thus increase the probability for obtaining the native-like structures for a given protein. Our second objective is to extend these structures to a full-atomic model in order to further approach the native-structure and to facilitate comparisons to assessment methods of other groups. All these developments should be especially useful for finding structures corresponding to sequences for which existing homologues have less than 20% identity. 2. Common realisations The web server front page has been done and is available to everybody. It might be interesting to put links to other web sites that members of the consortium estimate relevant. Nikolaos Papandreou will take in charge this gateway. It was previously decided to link our entries of the protein set to the PDB entries; instead it is much better to use PQS (Protein Quaternary Structure) at EBI as we are interested in the quaternary structures (one has to check if there is a problem of copyright). The logo is still waiting proposals. After discussion, we decided to change the format of the dataset that was decided in Paris. It appears that Fasta format is difficult to handle as long as structure is concerned, and we decided to adopt the DSSP format. Jean-Marc Kwasigroch will send within a couple of weeks, a test set of ten PDB files in DSSP format. There will be a certain number of colums already used, and each group will have to make its decision upon the number of columns they do need. This format will have to be definitly fixed at the Manchester meeting. 3. Discussion of the Work Packages We have split in three parts to be more efficient. Groups 1, 2, 3, 4 and 5 discussed about WP2, fragments and structures. Groups 5, 6 and 7 discussed of nucleus and structure, i.e. WP3, and groups 4 and 6 discussed about fingerprints of WP4. Groups 1, 2, 3 and 4 So far, on a reasonable data set, it seems a good correlation between topohydrophobic positions and MIR or between topohydrophobic positions and TEF. It will have to be extended to the full database. Correlation will have to be done between foldons and protofragments and TEF. We also will have to find a consensus which might be based on the best guess. Edward reported about work of his student, E. Aharonovsky, about vocabulary of three-letter words that statistically display preference to be at distance 25-30 residues one from another – presumably, the sequences of the locks closing the loops. The work is close to completion, and the words (about 200 of total 8000 triplets) soon will be open for the collaborating groups. Once we receive the 200 words from Edward, one has to check wether or not they map the ends of the TEF. Groups 5, 6 and 7 One has to discriminate the structures to find the native ones. One wishes to extend the database of a few entries (1ubq and 1cro, for instance). This has to be proposed fairly rapidly to Nikolaos. Natively non folding sequences might be interesting to study; although there is no available structure, there is some experimental information that one could use. The alignment viewers such as CINEMA and UTOPIA, freely available for the consortium, will have to be improved. The last version of Fold-X should be used. Actually group 4 will gather all the data from other groups and will see how they match to the fingerprints. 4. Next meeting It will be held in Manchester on May 2004. Terri will organise it and she will determine the date in the early days of January. It will be important for Manchester to start to produce pair wise correlation of the different methods. 5. People present at Bruxelles meeting This meeting was attended by the following people : - Jacques Chomilier and Anne Lopes from Group 1 - Nikolaos Papandreou and Elias Eliopoulos from Group 2 - Edward Trifonov from Group 3 - Therese Attwood and Manuel Corpas from Group 4 - Marianne Rooman, Christophe Biot, Yves Dehouck, Jean-Marc Kwasigroch, Dimitri Gilis and René Wintjens from Group 5 - Luis Serrano, Raphael Guerois, Frédéric Rousseau from Group 6 - David Perahia, Charles Robert and Liliane Mouawad from Group 7