Essential Bioinformatics and Biocomputing Module (LSM2104)
advertisement

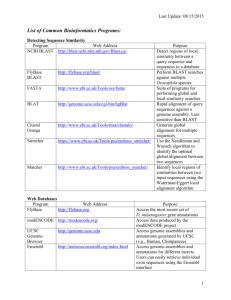
Sequence analysis Practical INTRODUCTION Hox proteins are involved in the regulation of development (morphogenesis) and have been identified in a diverse group of organisms, such as animals, fungi and plants. Hox proteins are known to contain a characteristic signature. The goals of this practical are to: 1. retrieve a protein from Genbank using an accession number. 2. Identify a homolog in another species using BLAST 3. Align the two sequence sequences using DOTMATCHER (http://sf01.bic.nus.edu.sg/EMBOSS/) 4. Identify homologs in additional species 5. Align all sequences using ClustalW 1. Extract the human Hox A4 (NP_002132) protein sequence in FASTA format from NCBI (http://www.ncbi.nlm.nih.gov/) and save it to a text file on your desktop. 2. Use the NCBI BLAST server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify homologs of Hox A4 in mouse, sea-bass (Morone saxatilis) and horn shark (Heterodontus francisci). Add these sequences to your protein text file Organism % identity between A4 orthologs E-values Human – Mouse Human – Sea-bass Human – Horn shark 3. Generate a dot-plot for human and shark Hox A4 proteins by aligning these sequences using the DOTMATCHER software of EMBOSS ((http://sf01.bic.nus.edu.sg/EMBOSS/) 3.1 How do you interpret the long diagonal line in the top right corner of the matrix? (remember to copy the dotplot into your report). 4. Using ClustalX, generate a multiple sequence alignment of all four Hox A4 proteins. 4.1 What region is conserved in all species …………………(amino acid position) 4.2 What is the corresponding exon number ……… 4.3 Save the alignment in a textfile. How is identity indicated in the alignment? 5. Align seq1 and seq2 using the DOTMATCHER software of EMBOSS ((http://sf01.bic.nus.edu.sg/EMBOSS/) 5.1 What scenario would explain the dot-plot pattern that you observe? 5.2 Modify the window size to 100. What is the effect on the dot-plot? >seq1 CACTCGATGCAGGCGCTGTCCTGGCGCAAGCTCTACTTAAGCCGCGCCAAGCT CAAAGCTTCCAGCCGGACCTCGGCTCTGCTCTCCGGCTTCGCCATGGTAGCGA TGGTGGAAGTCCAGCTGGACACAGACCATGACTACCCACCAGGGTTGCTCATC GTCTTTAGTGCCTGCACCACAGTGCTAGTGGCCGTGCACCTGTTTGCCCTCATG ATCAGCACCTGCATCCTGCCCAACATCGAGGCTGTGAGCAACGTCCACAACCT CAACTCGGTCAAAGAGTCACCCCACGAGCGCATGCATCGCCACATCGAGCTGG CCTGGGCCTTCTCCACGGTCATCGGGACGCT >seq2 CACTCGATGCAGGCGCTGTCCTGGCGCAAGCTCTACTTAAGCCGCGCCAAGCT CAAAGCTTCCAGCCGGACCTCGGCTCTGCTCTCCGGCTTCGCCATGGTAGCGA TGGTGGAAGTCCAGCTGGACACAGACCATGACTAATGATCAGCACCTGCATCC TGCCCAACATCGAGGCTGTGAGCAACGTCCACAACCTCAACTCGGTCAAAGAG TCACCCCACGAGCGCATGCATCGCCACATCGAGCTGGCCTGGGCCTTCTCCAC GGTCATCGGGACGCT 6. Questions about BLAST 6.1 What is a high scoring sequence pair (HSP)? 6.2 What is the Expect value? 6.3 Why is the following statement false: “The query length and query composition affects the Expect value”.